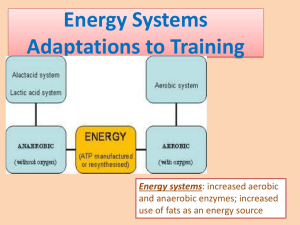
Full text
advertisement

A mathematical model for estimating
anaerobic synthesis rate constants for
Escherichia coli proteins.
Tijdens dit bachelorproject werd een wiskundig model ontwikkeld om de synthese constanten
van eiwitten in de bacterie Escherichia coli onder anaerobe omstandigheden te kunnen
schatten. Dit model gebruikt experimenteel verkregen waarden als startpunt van de
rekenmethode. De startwaarden zijn verkregen gedurende een pulse-chase experiment waarin
eiwitten werden gelabelled. Met behulp van moderne analytische technieken zijn deze
eiwitten geïdentificeerd en zijn een tweetal ratio's bepaald: de Rnew, welke het aantal nieuw
gesynthetiseerde eiwitten weergeeft en de Rtotaal, welke het totaal aantal eiwitten weergeeft.
Deze twee ratio's worden door het model als startpunt gebruikt.
Het model werd ontwikkeld met behulp van het computerprogramma MATLAB en gebruikt
diverse functies en algoritmes om differentiaalvergelijkingen op te lossen welke de Rnew en
de Rtotaal definiëren. Centraal in dit model staat de ∆R formule; deze formule beschrijft de
relatie tussen de gemeten ratio's van Rnew en Rtotaal en de voorspelde waarden welke door het
model worden bepaald. De ratio's zijn opgebouwd uit een drietal parameters; kdeg (de
degradatieconstante) ksyn,aerobic (de aerobe synthese constante) en de ksyn,anaerobic (de anaerobe
synthese constante. De eerste twee parameters zijn vooraf gedefinieerd waardoor alleen de
ksyn,anaerobic een variabele is.
Met behulp van het model kan deze laatste variabele worden geschat.
Wouter W. Woud (5974003)
Bio-Exact
4-07-2012
Supervisor: Prof. Dr. C. G. de Koster
Swammerdam Institute for Life Sciences
0
Index
Introduction...................................................................................................... Page 2
Experimental and Theoretical background...................................................... Page 4
The Model........................................................................................................ Page 9
Results.............................................................................................................. Page 16
Conclusion........................................................................................................ Page 21
APPENDIX…………………………………………………………………… Page 22
- The Source Code…………………………………………………... Page 22
- Proteomic Data Set……………………………………………….... Page 27
References.......................................................................................................... Page 31
1
Introduction
Many structural and functional properties of all living cells are defined by proteins. They are
responsible for executing many different tasks within the cells, as specified in the cell’s
DNA1. Because of this key functionality, proteins make up a large part of a cell’s
constituents. For example, half of the dry mass of an Escherichia coli cell is made up by
proteins. The other half consists mostly of DNA and RNA2.
Important insights about cellular functionality can be gained by studying protein activity and
interactions within the cell. As such, protein research is an active research field. Classic
methods of study mostly consist of protein isolation and/ or labelling. A number of
techniques have been developed to isolate proteins from cells. Most noticeably various types
of chromatography can be used to isolate proteins based on different properties such as
molecular weight, charge or binding affinity3. Labelling techniques such as the radioactive
isotope pulse-chase labelling with 35S-methionine or the use of Fluorescent Proteins (FP) can
be used to make proteins inside the cell visible under a microscope4,5. These techniques are
commonly used by scientists for various research purposes.
Protein isolation and labelling have helped to gain useful information about a wide range of
proteins. The information gained using these techniques mostly concerns protein structure or
functionality. Another part of the research field is more interested in the total ensemble of
proteins present in a cell at certain points in time. These studies generate huge data sets, and
form the field of proteomics, named after the related field of genomics, which handles large
data sets of genetic information. Important techniques in proteomics that are able to deal with
large amounts of proteins are 2D electrophoresis6 and mass-spectrometry7 .
Using 2D electrophoresis thousands of proteins can be separated simultaneously in one
separation procedure whereas mass-spectrometry allows proteome wide high-throughput
sequencing of peptides and identification of proteins. 2D electrophoresis enables the
separation of complex mixtures of proteins according to isoelectric point (pl), molecular mass
(Mr), solubility and relative abundance. Furthermore, it delivers a map of intact proteins
which reflects changes in protein expression level, isoforms or posttranslational
modifications.
This is in contrast to mass-spectrometry based methods which perform analysis on peptides
where Mr and pl information is lost and where stable isotope labelling is required for
quantitative analysis.
The huge amounts of data generated using these and other techniques find its application in
the computational systems biology. This relatively new field aims to develop and use
efficient algorithms, data structures and visualization and communication tools with the goal
of computational modeling of biological systems8 . More specifically, protein data acquired
through, for example, mass-spectrometry of whole cells can be used to create computer
models that can predict protein characteristics in these cells. Characteristics of interest may
include rates of protein synthesis and degradation and related to the latter the protein half-life,
which is the time it takes for half of a given protein population to degrade. When compared to
experimental data an attempt can be made to link these predicted characteristics to protein
functionality in the living cell.
The basis of such a predictive computer model is formed by a set of mathematical equations
that describe the behavior of proteins in the organism of interest. To simplify these equations
certain assumption concerning this behavior can, and sometimes must be made, as is also
shown later in this paper. Arguments for such assumption must be obtained from
experimental research. By optimizing model parameters such as reaction rate constants to
2
match experimental results a valid descriptive and predictive model can be created for the
biological system of interest.
In the case of whole cell proteomics an ideal model would for example be able to estimate
both synthesis and degradation constants as well as half-lives for a variety of proteins. If
experimental research provides errors for measurements, e.g. standard deviations, the model
should be able to process these as well when estimating synthesis and degradation constants.
Ultimately this model should be usable to gain useful insights in the proteome dynamics of
the organism of interest.
3
Experimental and theoretical background
The aim of our research is to build a mathematical model which can be used to estimate
reaction rate constants of both synthesis and degradation as well as half-lifes of given
proteins. Our mathematical model has been created using the computer program MatLab®
and uses a proteomic data set as input. This proteomic data set was generated during a pulsechase labelling experiment using the methionine analog azidohomoalanine (abbreviated as
‘azhal’)9. Whilst our model can be used to estimate both reaction rate constants and half-lifes,
this paper will only discuss the part of the model which estimates the anaerobic reaction rate
constants.
The proteomic data set was generated during a pulse-chase labelling experiment. This
experiment was used to identify and quantify several hundreds of newly synthesized proteins
in Escherichia coli upon pulse labelling with the methionine analogue azidohomoalanine
(azhal). For the first 30 minutes after inoculation, a methionine-auxotrophic E. coli strain
grows equally well on azhal as on methionine under aerobic conditions. Upon a pulse of 10
minutes with the methionine analogue azidohomoalanine (azhal), a change in environmental
conditions by switching from aerobic to anaerobic circumstances and digestion of total
protein, newly synthesized azhal labelled peptides are isolated by a retention time shift
between two reversed chromatographic runs. The retention time shift is induced by a reaction
selective for the azido group in labelled peptides using tris(2-carboxyethyl)phosphine.
Selectively modified peptides are identified by reversed phase liquid chromatography and
online tandem in mass spectrometry. As methionine is the more dominant competitor over
azhal, the medium should be purged of methionine before labelling for maximum
incorporation10. Thus, by using this pulse-chase experimental setup, newly synthesized
proteins can be identified because of their incorporation of the methionine analogue azhal.
Furthermore, this strategy allows us to identify changes in total protein levels on the same
time scale as new protein synthesis. Using these values for total protein level and newly
synthesized proteins two different ratios were obtained. These ratios will be explained shortly.
Model
During the exponential growth phase of E. coli cells, we assume that the number of proteins
increases proportional to cellular mass and that protein synthesis and degradation are firstorder with the number of proteins.
At the beginning of the experiment (t = 0), environmental conditions are altered from aerobic
to anaerobic and only non-labeled proteins are present in E. coli. These unlabelled proteins
are named Pold as they solely exist as pre-existing material, and thus only methionine
containing peptides are present. After 10 minutes of Azhal labelling under anaerobic
conditions the unlabelled proteins have been partly degraded and new proteins which contain
the azhal label have been synthesized. These labeled proteins are named Pnew. The
degradation of Pold is given by the following differential equation:
4
Pold
k deg * Pold (1)
t
We assume that protein synthesis is of the first-order with the total amount of protein present
in the cell. Therefore, Pnew is being formed with a first-order rate constant ksyn and degraded
by kdeg. We assume that kdeg has the same value for both labelled and unlabelled proteins, as
well as for aerobic and anaerobic conditions. Arguments for this assumption can be found in
the supplementary data of the publication ‘Proteome-wide alterations in Escherichia coli
translation rates upon anaerobiosis’, Kramer et al9.
As such, the rate of change of azhal containing peptides (Pnew) is defined by the following
differential equation:
Pnew
k syn * Pold (k syn k deg ) Pnew
t
( 2)
The term ksyn*Pold seems a little strange at first, but recall that we assume that protein
synthesis is of the first-order with the total amount of protein present in the cell. Therefore,
the concentration of newly formed proteins depends on both the pre-existing amount of
protein (unlabelled, Pold) and the newly synthesized amount of protein (labelled, Pnew).
Integration of both differential equations is straightforward and yields the time-dependant
functions Pold(t) and Pnew(t):
Pold (t ) Pold (0) * e
kdegt
Pnew (t ) Pold (0) * [e
(3)
( k syn kdeg ) t
e
kdegt
] (4)
After summation of Pold(t) and Pnew(t) the term Ptotal(t) is acquired. This term reflects the total
amount of protein present at any given time and is defined by the following equation:
Ptotal (t ) Pold Pnew Pold (0) * e
( ksyn kdeg ) t
(5)
As described earlier our model uses data derived from a proteomic data set as input. This
experimentally acquired data is being used to create two different ratio’s named Rnew,measured
and Rtotal,measured which are consequently being used as input entry in our model.
How exactly are the ratios of Rnew and Rtotal defined? The first ratio is the number of newly
synthesized proteins during the pulse labelling time interval upon transition from aerobic to
anaerobic conditions. The ratio of newly synthesized proteins (Rnew) under anaerobiosis (Pnew,
anaerobic) and aerobiosis (Pnew, aerobic) is defined as:
Rnew
Pnew, anaerobic(t )
Pnew, aerobic (t )
Pold , anaerobic(0) * [e
Pold , aerobic (0) * [e
5
( k syn, anaerobic k deg ) t
( k syn, aerobic k deg ) t
e
e
k deg t
k deg t
]
]
(6)
Please note that these parameters strongly represent equation 4. Equation 7 can be reduced to
an easier form by replacing the subtractions made with -1, thus obtaining equation 8. This
substitution can be made as in both the dividend and the divisor the same value is being
subtracted.
Pold , anaerobic(0) e ksyn, anaerobic t 1
Rnew
*
(7 )
Pold , aerobic(0) e ksyn, aerobic t 1
The second ratio is the copy number of total protein level at the end of the labelling time
between the two environmental conditions. This ratio is derived from the ratios of peptides
that do not contain methionine or azhal. Azhal peptides are excluded since these represent
exclusively newly synthesized material, while methionine peptides are excluded because they
represent only pre-existing material. The non azhal/methionine-containing peptide copy
number equals in the kinetic model the summation of Pold(t) and Pnew(t), as described in
equation 5.
(k
k ) t
( Pold (t ) Pnew (t )) anaerobic Pold , anaerobic(0) e syn, anaerobic deg
Rtotal
*
(8)
( Pold (t ) Pnew (t )) aerobic
Pold , aerobic (0) e ( ksyn, aerobic kdeg )t
This total protein ratio reflects the overall protein expression between the two environmental
states. In our model these two experimental ratios (Rnew & Rtotal) are predicted by three
different reaction rate constants (ksyn, anaerobic, ksyn, aerobic and kdeg). To compare the predicted
ratios of Rnew and Rtotal with the measured ratios (as is the ultimate goal, as can be seen in
equation 13) several assumptions are made. First of all, as stated before the aerobic culture is
in the exponential growth phase. Thus, the whole of metabolic, proteomic and genomic
activities within the E. coli cells are set for reproduction. When the environmental conditions
are altered (form aerobic to anaerobic) a growth arrest of approximately 10 minutes occurs
within the E. coli cells. During these 10 minutes, the E. coli cells are altering their
transcriptome and proteome in response to the changed environmental conditions.
Secondly, to estimate protein half-lifes from the proteomics data set the value of kdeg is being
varied. More information on how this variation in kdeg occurs can be found in “ A
mathematical model for estimating protein half-lifes in Eschericia coli” by Tom Boer.
From literature it is known that there are approximately two different protein populations
with regard to protein degradation rates within E. coli: a small rapidly degrading population
with half-lifes lower than 10 minutes and a more slowly degrading population with half-lifes
ranging between several hours up to 23 hours11,12. The degradation rate constant kdeg is
inversely connected to protein half-life by the following equation:
k deg
ln 2
(9)
As kdeg has been defined, the next step would be the estimation of ksyn, aerobiosis. During the ten
minutes labelling time (tp) the optical density (O.D.600 nm) of the E. coli culture increases with
a factor P(tp) divided by P(t0). This implies that biomass increases with the same factor and
6
using the assumption that total protein content scales linearly with biomass the increase of
protein mass also equals P(tp) divided by P(t0). Furthermore, the relative protein composition
does not change during steady state aerobic growth. This means that the increase in copy
number for each protein is also P(tp) divided by P(t0). For each protein, ksyn, aerobiosis is related
to its kdeg during steady state aerobic conditions via:
k syn,aerobiosis
1 P(t p )
ln
k deg r k deg (10)
t p P (t 0 )
And is related to protein half-life as:
k syn,aerobiosis r
ln 2
(11)
The E. coli cells used to generate the data set were exponentially growing and P(tp)/P(t0)
yielded a value of 1.0625. This value was named ‘growth’ in our model. The ‘r’ value as
notated in the equations above was calculated using the pulse time and the growth value, as
described in equation 13. Using this equation, the ‘r’ value thus yields a fixed value of 0.0026.
Note that the ‘r’ value is the same for all proteins.
r
1
* log( growth) (12)
pulsetime
Thus, all of the parameters needed to calculate values of Rnew,calculated and Rtotal,calculated are now
defined. Please notice the difference between Rnew,measured / Rtotal,measured and Rnew,calculated /
Rtotal,calculated, the first being the ratio’s our model uses as input from the proteomic data set
and the later being the ratio’s which will be used to estimate the anaerobic synthesis constant
ksyn,anaerobic.
The anaerobic synthesis constant is being estimated by using the following formula:
R Rnew,calculated Rnew,measured Rtotal,calculated Rtotal,measured
(13)
Equation 13 can be seen as the summation of the absolute values of a variable minus a fixed
value. As both Rnew, measured and Rtotal,measured are fixed values (inputted directly from the
proteomic data set) and ∆R is being minimized as much as possible to ascertain our model
matches reality as much as possible, we are able to find a value of ksyn,anaerobic.
For example, let's set ∆R = 0, and use the Rnew, measured and Rtotal,measured values of aldehydealcohol dehydrogenase (ADHE) from the proteomic data set. When both Rnew,calculated and
Rtotal,calculated would be written as equations 7 and 8, equation 13 would look like this:
Pold , anaerobic(0) e ksyn, anaerobic t 1
Pold , anaerobic(0) e( ksyn, anaerobic kdeg )t
0
* ksyn, aerobic t
10,26
* ( ksyn, aerobic kdeg )t 2,59
Pold , aerobic(0) e
Pold , aerobic(0) e
1
7
(14)
As stated before, Rnew,calculated and Rtotal,calculated consist of 3 different parameters: kdeg, ksyn,aerobic
and ksyn,anaerobic. As both kdeg and ksyn,aerobic have been defined only one parameter is subject to
change: the ksyn,anaerobic.
Thus, the Rnew,calculated and Rtotal,calculated values can be used, in combination with the
minimization of ∆R, to obtain values for ksyn,anaerobic. How the estimation of ksyn,anaerobic occurs
exactly will be explained in great detail in chapter 3, ‘The Model’.
8
The Model
Our model runs in various stages, which are presented schematically in figure 1. The first
step is the extraction of data from the proteomic data set. Secondly, normally distributed
random numbers are generated over the extracted data. After those numbers have been
generated, a set of 4 differential equations will be solved in combination with a least square
function called lsqnonlin. The last steps are the calculation of the anaerobic synthesis
constant ksyn,anaerobic and the' re-writing' of the calculated data into the proteomic data set.
Figure 1: A schematic overview of the model.
The various steps of the model described above will be discussed in great detail within this
chapter. We begin this chapter with an introduction to the differential equations.
Our model uses a set of 4 differential equations in order to estimate the anaerobic synthesis
constants ksyn,anaerobic from E. coli proteins. These four different differential equations
(equation 14) represent the amount of unlabelled protein present under aerobic conditions
(differential equation 1, Pold, aerobic), the amount of labelled protein present under aerobic
conditions (differential equation 2, Pnew,aerobic), the amount of unlabelled protein present under
anaerobic conditions (differential equation 3, Pold,anaerobic) and the amount of labelled protein
present under anaerobic conditions (differential equation 4, Pnew,anaerobic). As we can see these
differential equations show strong similarities with equations 1 & 2 from the previous chapter.
9
dy1
dt
dy 2
dt
dy3
dt
dy 4
dt
k deg *
y1
dt
k syn,aerobic *
k deg *
y1
y2
(k syn,aerobic k deg ) *
dt
dt
y3
dt
k syn,anaerobic *
y3
y4
(k syn,anaerobic k deg ) *
dt
dt
Paerobic
y1 y 2
Panaerobic
y3 y 4
y4
(15)
y2
P
anaerobic
Paerobic
Rnew, calculated
Rtotal, calculated
The equations described above (equation 14) are being combined in the same ways as
described in the previous chapter (equation 5). By adding dy1 and dy2 a new equation can be
dt
dt
obtained which represents the total amount of protein present under aerobic conditions. This
adding process can be repeated for dy3 and
dt
dy4 which
dt
returns a function representing the total
amount of protein present under anaerobic conditions. By dividing equation 4 with equation 2
the parameter Rnew, calculated is being obtained. Rtotal,calculated is being obtained by dividing the
total amount of protein present under anaerobic conditions (Panaerobic) through the total
amount of protein present under aerobic conditions (Paerobic).
As stated before, the aim of our research is to build a mathematical model which can estimate
the anaerobic synthesis constants ksyn,anaerobic of various proteins from E. coli. For this purpose
a mathematical model was build using the computer program MatLab®, the MathWorks. This
model heavily relies on the series of equations described thus far, and will be discussed in
great detail within this chapter. Both the source code of the model and the excel file
containing the Rnew,measured and Rtotal,measured values derived from the proteomic data set can be
found in the appendix ‘Proteomic Data Set’, page 26.
Data extraction
Our model starts with the extraction of the Rnew, measured and Rtotal,measured values from the
proteomic data set. The data in the first column of the proteomic data set represents the
protein number, which allows us to easily find the number of proteins our model will use
when operating. The second column represents the protein acesion number, a code with wich
the protein can ben identified. The third column represents the protein names, e.g. ADHE for
aldehyde-alcohol dehydrogenase. The fourth, fith, sixth and seventh columns of the
proteomic data set represent the measured values of Rnew and Rtotal and their corresponding
standarddeviations. The other columns represent the outputted data; more information
provided at the end of this chapter.
The data extraction has been automated in such way that when running the model a pop-up
screen appears which will ask the user to select an excel file of choice. The codes for this
feature are shown below:
10
% read excel data
[filename, pathname] = uigetfile('*.*','Select data file');
path = [pathname,filename];
[num, txt, raw] = xlsread(path);
[lcol, lrow] = size(num);
Matlab automatically separates text and numerical values from the extracted data. Numerical
values are returned in the array num and text will be returned in the array txt. If a cell
contains both numerical values and text it is considered as raw data and will be stored in the
array raw . This feature is extremely handy as it solves the problem of separating text and
numerical values manually and thus saves a lot of time. Also, the size of the numerical array
is being determined. As our proteomic data set has its proteins listed from top to bottom, the
array lcol is a direct indication of the amount of proteins extracted from the excel file.
However, as can be seen in the appendix 'Proteomic Dataset' our model does not use all
numerical values provided by the excel file: the first numerical value encountered, the
'protein number', has no value for the estimation of the anaerobic synthesis constants. Only
the values of Rnew, measured and Rtotal, measured and their corresponding standard deviations are
needed for our calculations. The standard deviations have been obtained by conducting the
pulse-chase experiment several times.
Creating normally distributed values of Rnew & Rtotal
The standard deviations are being used by the following code line to generate a number of
normally distributed values of Rnew,measured and Rtotal,measured around the given means of both
values.
% create n normally distributed randoms around Rnew
randoms(:,1) = random('norm',num(g,4),num(g,5),[n 1]);
% create n normally distributed randoms around Rtotal
randoms(:,2) = random('norm',num(g,6),num(g,7),[n 1]);
These codes have a two folded functional character and operate as follows: the first position
after random is specified for the type of distribution one would like to create. In our model,
we wanted the values of Rnew,measured and Rtotal,measured to be distributed normally and therefore
the options have been set to 'norm'. The second and third positions specify the mean and
standard deviation respectively. As can be seen, these values are being extracted from the num
array which was created previously. For example, the code num(g,4)implies that the
numerical value of row g and colomn 4 are being used as input for the mean value of
Rnew,measured. Since every g represents a protein in our model this code ensures that normally
distributed randomly generated values of both Rnew, measured and Rtotal, measured are being created
for all proteins.
The last position, [n 1], determines the size of the array in which the randomly created
values of Rnew,measured and Rtotal,measured are being stored. This array has the size of a matrix with
n rows and 1 column. Because the n parameter has been set to a value of 300, a [300 1]
matrix will be created, thus resulting in 300 randomly generated normally distributed values
of both Rnew,measured and Rtotal,measured. Since the n parameter has been defined at the start of the
model (appendix: ‘The Source Code’) the number of randoms generated can easily be
adjusted by changing the value of this parameter.
The two folded functional character of these codes are due to the fact that they both generate
random numbers as well as extract the correct data from the num arrays created by the
11
xlsread
function.
The random numbers generated are stored in the arrays Rnewmeasured and Rtotalmeasured
by the following command line:
% optimize ksynanaerobic for all randoms
for p = 1:n
Rnewmeasured = randoms(p,1);
Rtotalmeasured = randoms(p,2);
Note that the random values have changed from an n index to a p index.
Calculation of ksyn,anaerobic
The randomly generated Rnew, measured and Rtotal,measured values are being used as input entry for
the ∆R formula (equation 13, chapter 2 ‘Experimental and theoretical background’). Equation
13 can be seen as the summation of the absolute values of a variable minus a fixed value.
That is, every randomly generated value for Rnew, measured and Rtotal,measured represents a fixed
value. In the specific case of the protein aldehyde-alcohol dehydrogenase (ADHE), the first
protein in the proteomic data set, equation 13 can be written in the following form:
R ADHE Rnew,calculated 10,26 Rtotal,calculated 2,59
(16)
Please note that the values of Rnew, measured and Rtotal,measured in this formula represent the
standard values as can be found in the appendix ‘Proteomic Data Set’, and not values which
have been randomly generated.
The parameters Rnew,calculated and Rtotal,calculated consist of three variables from which two have
been defined previously (ksyn,aerobic and kdeg), resulting in only one variable left for estimation.
This last variable, ksyn,anaerobic (denoted x in our model), is being estimated by Matlab using
an optimization routine function called ‘lsqnonlin’. This function solves nonlinear leastsquares problems, including nonlinear data-fitting problems. The function lsqnonlin in our
model is being called on by the following command lines:
[x,resnorm] = lsqnonlin(@diff,x0,lb,ub,options);
options = optimset
('Display','off','Largescale','off','Algorithm','levenberg-marquardt');
The function lsqnonlin is designed, as stated before, to solve non-linear least-square
problems. Since the ∆R value has to be as small as possible for our model to reflect reality,
our model faces a non-linear least square problem and thus the lsqnonlin function is being
used.
However, before the operation of the lsqnonlin function can be explained we first have to
address the problem of solving the differential equations as described in equation 15. If we
take a closer look at the lsqnonlin command line one can see that the function tries to find a
minimum in the sum of squares of the functions described in diff. Its starting value for x
(denoted as x0) is set to 0, and both the upper and lower bounds are empty (lb = [] & ub
= []) implying no limits for lsqnonlin to find values of x which result in a minimum of ∆R.
These values for x0,lb,ub are set at the first section of the source code (% define
lsqnonlin parameters).
12
The command lines for the diff function are depicted below. As one can see the diff
section is a rather large part in our model, and will therefore be discussed step by step.
function deltaR = diff(x)
% solve the ode as described in the function 'kinetics'
options = odeset('RelTol', 1e-4);
[t,sol] = ode45(@kinetics,tspan,y0,options,x);
The first step within the diff function is solving the differential equations described in
equation 14. This solving process is carried out by the function ode45, a solver that integrates
ordinary differential equations. The differential equations which are to be solved are situated
in the function kinetics (@kinetics), which can be found at the end of the source code. The
interval of integration is set by tspan and equals the duration of the pulse-chase experiment,
which was ten minutes in our model. Thus, tspan = [0 pulsetime]. The starting values for
the differential equations are set by y0 and are 100, 0, 106.25 and 0 respectively. Recall that
the first and third differential equations represent the amount of unlabelled protein present
under aerobic conditions and the amount of unlabelled protein present under anaerobic
conditions respectively, and thus will only decrease over time. The options only contain a
function that controls the number of digits in all solution components, assuring all outputted
entries have the same length. The only variable in here is x, and all solutions are stored in a
matrix of size [t,sol].
While using this solving routine, problems were encountered in the form of NaN errors.
This problem was solved by removing the solving values at t=0 from the [t,sol] matrices.
% remove y0 from data to prevent NaN errors
sol(1,:)=[];
The next step is to extract the y vectors from the matrices and to assure all vectors have the
same length. These objectives are achieved by the following command lines:
% extract y vectors from the sol matrix
y1 = sol(:,1);
y2 = sol(:,2);
y3 = sol(:,3);
y4 = sol(:,4);
% fix
ny1 =
ny2 =
ny3 =
ny4 =
y vector length to prevent matrix dimension errors
y1(1:z);
y2(1:z);
y3(1:z);
y4(1:z);
The vector lengths are being set by parameter z, which is set to the value of 56. This number
was empirically found the highest value for which our model would operate without any
errors (vector length mismatch).
The last step occurring in diff is the actual calculation of ∆R. The values calculated by the
ode45 function are being combined in the same ways as described in equation 14, resulting in
values of Razhal and Rtotal, also known as Rnew,calculated and Rtotal, calculated. By using
equation 13 our model now is able to calculate ∆R.
13
% calculate the difference between measured en predicted data
Paeroob = ny1 + ny2;
Panaeroob = ny3 + ny4;
Razhal = ny4 ./ ny2;
Rtotal = Panaeroob ./ Paeroob;
deltaR = abs(Razhal- Rnewmeasured)+abs(Rtotal-Rtotalmeasured);
deltaR = deltaR(z);
end
Since we are only interested in the final value of ∆R (that is, the value of ∆R after ten minutes
of labelling), the 56th entry is being taken as the final value of ∆R (deltaR = deltaR(z)).
Thus, the function lsqnonlin solves the differential equations in such way that the ∆R value
becomes a minimum, making our model reflect reality as good as possible.
The anaerobic synthesis constant ksyn,anaerobic is the x value for which the minimum is achieved,
and is being extracted by the following command line:
ksynanaerobic(p) = x;
Please note the p index and recall that the values of Rnew,measured and Rtotal,measured in reality
consist of 300 randomly generated normally distributed values. This means that there are 300
different values of ksyn,anaerobic as well. The final value of ksyn,anaerobic as well as its standard
deviation are being calculated by taking the mean of all 300 values of ksyn,anaerobic and using
n
the formula n
i 1
( xi ) 2
respectively.
n
These calculations are being achieved by the following codes:
ksynanaer(g) = mean(ksynanaerobic);
ksynanaersd(g) = sqrt((sum((ksynanaerobic-mean(ksynanaerobic)).^2))/n);
Returning data to excel file
The final step in our model is the transfer of the calculated data back to the proteomic dataset.
This is being achieved by the following command lines:
% write data to Excel
disp('Writing...')
xlswrite(path, {'ksynanaer'}, 1, 'K1')
xlswrite(path, ksynanaer', 1, 'K2')
xlswrite(path, {'sd'}, 1, 'L1')
xlswrite(path, ksynanaersd', 1, 'L2')
Recall that during the data extraction, the first step in our model, an excel file was chosen and
stored in the array path. The code lines described above use this same path (first entry after
xlswrite) to write both the ksyn,anaerobic and their standard deviations into the proteomic data
set. The second entry defines the outputted data, and the third and fourth entries define the
sheet and range respectively. Note that the second entry contains either {'ksynanaer'} /
{'sd'} or ksynanaer' / ksynanaersd'; the first one (purple) can be seen as a title for the
outputted data whereas the second line represents the actual matrix containing the calculated
data.
14
Additional features
Our model uses a set of codes to help us monitor its calculation progress. This set of codes
represents a timer function, enabling us to monitor the time our model needed to complete its
calculation procedure. These codes use the tic/toc combination and are depicted below:
tic
‘Calculation Zone’
Elapsedtime = toc/60;
str = ['Elapsed time: ', num2str(Elapsedtime), ' minutes'];
disp(str)
The tic code starts a stopwatch timer which runs in seconds, and the toc code stops this
timer. The tic code should be placed at the very start of the model, that is, just before the
model takes its first action. As the first action of our model is the extraction of data from the
proteomic data set, tic should be placed just before the % read excel data
codes.
Just as tic should be placed at the start of the model, toc should be placed at the end of it.
As our model ends with the transfer of the calculated data back to the proteomic data set, toc
should be placed right after these codes. Thus, the ‘Calculation Zone’ represents all the
function and codes in our model which play a role in estimating the anaerobic synthesis
constants. In the codes above, the value of toc is being divided by 60 to acquire the time in
minutes. The calculation time then is being displayed by disp(str), which displays ‘Elapsed
time: (calculation time) minutes’.
15
Results
Effect of the randoms on the ksyn,anaerobic
In order to describe the effect of the number of normally distributed Rnew and Rtotal values
generated by our program on the estimation of ksyn, anaerobic and to find out what the minimal
value of n required is, a computational experiment was done by running our model with a
varying number of randoms whilst estimating the anaerobic synthesis rate constant for 1
protein only. The protein we selected for this experiment was aldehyde-alcohol
dehydrogenase (ADHE).
We ran our model using n = 1, 50, 100, 150, 200, 250 and 300 numbers of randoms over the
values of Rnew, measured and Rtotal, measured. A total of ten simulations for each random number
value (n) were done, and the outcomes of the values of ksyn,anaerobic were plotted against the
number of randoms used to calculate them. Note, the ksyn,anaerobic values calculated this way are
avaraged; e.g. generating 300 values of Rnew,measured and Rtotal,measured still lead to one value of
ksyn,anaerobic per simulation. The results of this experiment are shown in figure 2.
Figure 2: Effect of the n value on the ksyn, anaerobic
Obviously, using only 1 random number leads to a wide variety in ksyn, anaerobic values (values
ranging between 0.087 and 0.178), whereas n values ranging from 50 to 300 all lead to a
very uniform distribution of ksyn, anaerobic values. Thus, the minimum number of randoms
needed to obtain an accurate value of ksyn, anaerobic could be set to 50.
However, if we exclude the date from n =1 and take the mean of all estimated ksyn,anaerobic
values per n, we obtain a new graph which shows the variation in ksyn, anaerobic for the n values
from 50 to 300 in great detail (figure 3). The error bars represent the standard deviation
within the meaned estimated values of ksyn,anaerobic and do not represent the standard
deviations calculated by our model.
16
Figure 3: The effect of the n value on the ksyn, anaerobic, meaned values.
This new graph shows us that the higher the n value used, the smaller the variation in ksyn,
anaerobic becomes; n values exceeding 300 will probably lead to even further narrowing of the
variation. However, we found that the higher the n value becomes the longer it takes for our
model to complete its calculations.
Calculation Time
To see whether we could compose a formula which could estimate the calculation time of our
model in advance we ran our model for the ADHE protein with n values ranging from 1 to
400, intervals of 20 and recorded the time needed for our model to complete its calculations.
The results of this experiment are depicted below.
Figure 4: The effect of the n value on calculation time, 1 protein calculated.
17
As we see the time needed for our model to complete its calculation procedure scales linearly
with the number of randoms used. To gain insight in how the calculation time scales with an
increase of proteins, the experiment was repeated for 2 proteins. Results of this experiment
are depicted below:
Figure 5: The effect of the n value on calculation time, 2 proteins calculated.
As we see the time needed for our model to complete its calculation procedure scales linearly
with the number of randoms used, regardless whether only 1 protein is being estimated or 2.
By comparing both trend line formulas we observe that the trend line formula for 2 proteins is
roughly twice as large as the trend line formula for 1 protein. With this information we were
able to compose a formula which roughly estimates calculation time in advance:
(t ) g * (0.0049 * n) (1.0018 * g )
In this formula, t stands for time in minutes, g stands for the number of proteins used and n
stands for the number of randoms used. To verify whether this formula also holds for 3 or
more proteins, we conducted a series of runs with our model. In these runs, both the number
of proteins (g) and the number of randoms (n) were being varied:
Parameter settings
Estimated Calculation Time
True Calculation Time
g=5
& n = 250
11,34 minutes
10,81 minutes
g=8
& n = 140
13,50 minutes
12,81 minutes
g=3
& n = 360
8,29 minutes
7,75 minutes
g = 10
& n = 80
13,94 minutes
13,49 minutes
g = 15
& n = 120
23,85 minutes
23,06 minutes
18
Tabel 1: Estimated Calculation Time compared with True Calculation Time.
Note: Parameter values chosen at random.
We conclude that our formula is able to roughly estimate the calculation time needed by our
model.
Verifying the distribution of the randomly generated values
So far we have discussed the effect of the number of randoms on the estimation of ksyn,anaerobic
and its calculation time, but it is still unknown whether all estimated values of ksyn, anaerobic by
fitting of our kinetic model are normally distributed.
To verify whether these values are distributed normally, a simulation for ADHE was done
using an n value of 10.000 (calculation time: over 50 minutes). After the simulation was
complete, we plotted all 10.000 different values of ksyn, anaerobic in a histogram using the
following order of functions:
x = min(ksynanaeroob):.005:max(ksynanaeroob);
hist(ksynanaeroob,x)
title('Random distribution of Ksyn,anaerobic for ADHE')
The first line specifies the location on the x-axis and the number of the bins by taking the
lowest value and the highest value out of the 10.000 estimated ksyn, anaerobic values and divides
that space in steps of 0.005. So the 1st bin in the histogram has the lowest ksyn,anaerobic value on
the left side, and the right side of this bin basically is the minimum value + 0.005. So the
width of every bin is 0.005. The second line creates the histogram by using the 10.000 values
of ksyn, anaerobic and the value set by ‘x’ (thus over how many bins the 10.000 ksyn,anaerobic data
should be distributed). The third line obviously adds a title to the graph. The results of this
simulation can be found in figure 6.
Random distribution of Ksyn,anaerobic for ADHE
1000
900
Bin Count: 926
800
Bin Center: 0.133
Bin Edges: [0.13, 0.135]
700
600
500
400
300
200
100
0
-0.05
0
0.05
0.1
0.15
0.2
Figure 6: Random distribution of ksyn,anaerobic for alcohol-aldehyde
dehydrogenase (ADHE)
19
As we created random values for Rnew,measured and Rtotal,measured by using a normal distribution
(chapter 3: ‘The Model’, Creating normally distributed values of Rnew,measured and Rtotal,measured)
we expected a curve that would look like a Gaussian distribution / bell shaped curve.
However, figure 6 seems to be skewed to the left and thus does not have the characteristics of
a normal distribution. Also, the mean of the distribution in figure 6 is situated at 0.133,
whereas the calculated mean, that is, the mean outputted by our model, has a value of 0.126.
This skewness can be explained due to the fact that the randomly generated values of
Rnew,measured and Rtotal,measured enter the differential equations as depicted in equation 15. Since
these differential equations are not linear, skewness may appear.
The Proteomic Data Set
The complete list of all estimated ksyn,anaerobic values and corresponding standard deviations
per protein can be found in the appendix ‘Proteomic Data Set’. Please note that both the
anaerobic synthesis constants as well as the half-lifes are presented within the proteomic data
set. For information on how the half-life values were obtained please read the paper “A
mathematical model for estimating protein half-lifes in Eschericia coli” by Tom Boer.
20
Conclusion
The aim of our project was to build a model which could predict values of anaerobic
synthesis rate constants of proteins in E. coli under anaerobic conditions. In order to achieve
this goal the software program MATLAB was used as described in the previous chapters, and
a list of anaerobic synthesis constants (denoted as ksyn, anaerobic) and their corresponding
standard deviation was outputted. This complete list can be found in the appendix (Proteomic
Data Set).
In order to output reasonably accurate values an n-value ranging between 50 and 300 should
be used. All values in the appendix were calculated using an n-value of 300. Using n-values
exceeding 300 will lead to more narrowed-down values for the anaerobic synthesis constants.
However, this will lead to an increase in calculation time.
While our model does output values for ksyn, anaerobic we cannot state that the values which are
being outputted are correct as referential material is scarce.
We believe our model can be used to calculate synthesis constants for other organisms as
well as for aerobic conditions by using different sets of differential equations and by
adjusting the Azhal pulse labelling experiment.
21
APPENDIX
The Source Code
function [HL,HLsd,ksynanaer,ksynanaersd] = omegatau
% define differential equation parameters
pulsetime = 10;
growth = 1.0625;
% define lsqnonlin parameters
x0 = 0;
lb = [];
ub = [];
% define ode solver parameters
y0 = [100; 0; 106.25; 0];
% read excel data
[filename, pathname] = uigetfile('*.*','Select data file');
path = [pathname,filename];
%#ok<NASGU> suppressed 'unused variable' underline.
[num, txt, raw] = xlsread(path);
%#ok<NASGU>
[lcol, lrow] = size(num);
%initiate program runtime timer
tic
disp('Reading...')
% define randoms and ode restriction parameters
n = 300;
z = 56;
% preallocate matrices for speed and cleaner m-file
HL = zeros(1,lcol);
HLsd = zeros(1,lcol);
ksynanaer = zeros(1,lcol);
ksynanaersd = zeros(1,lcol);
ksynanaerobic = zeros(1,n);
deltaR4 = zeros(lcol,11);
deltaR5 = zeros(lcol,n);
% initiate progressbar
progressbar('Protein','Randoms','Total HL progress','Calculating HL');
for g = 1:lcol
Rnewmeasured = num(g,4);
Rtotalmeasured = num(g,6);
% progressbar parameter
22
b=1;
% HL optimum is narrowed down using intervals to reduce calculation time
hlrange = 1:100:10001;
deltaR = calcdeltaR;
[minvalue,position] = min(deltaR);
HLtemp = hlrange(position);
hlright = HLtemp+100;
hlleft = HLtemp-100;
if hlleft <= 0
hlleft = 1;
hlright = hlright+1;
end
b=2;
hlrange = hlleft:10:hlright;
deltaR = calcdeltaR;
[minvalue,position] = min(deltaR);
HLtemp = hlrange(position);
hlright = HLtemp+10;
hlleft = HLtemp-10;
if hlleft <= 0
hlleft = 1;
end
b=3;
% run final 20 HL's to find optimal HL
hlrange = hlleft:hlright;
deltaR = calcdeltaR;
[minvalue,position] = min(deltaR);
HL(g) = hlrange(position);
p = 0.5;
% add a decimal point to HL
for h = (HL(g)-p):0.1:(HL(g)+p)
halflife = h;
r = (1 / pulsetime)*log(growth);
kdeg = log(2) / halflife;
ksyn = r + kdeg;
options =
optimset('Display','off','Largescale','off','Algorithm','levenbergmarquardt');
[x,resnorm] = lsqnonlin(@diff,x0,lb,ub,options);
i = (HL(g)-p):0.1:(HL(g)+p);
[a,b] = size(i);
fv = find(i==h);
deltaR4(g,fv) = diff(x);
23
progressbar([],[],3/4,fv/b);
end
[minvalue,position] = min(deltaR4(g,:));
HL(g) = i(position);
halflife = HL(g);
r = (1 / pulsetime)*log(growth);
kdeg = log(2) / halflife;
ksyn = r + kdeg;
% create n normally distributed randoms around Rnew
randoms(:,1) = random('norm',num(g,4),num(g,5),[n 1]);
% create n normally distributed randoms around Rtotal
randoms(:,2) = random('norm',num(g,6),num(g,7),[n 1]);
% optimize ksynanaerobic for all randoms
for p = 1:n
Rnewmeasured = randoms(p,1);
Rtotalmeasured = randoms(p,2);
options =
optimset('Display','off','Largescale','off','Algorithm','levenbergmarquardt');
[x,resnorm] = lsqnonlin(@diff,x0,lb,ub,options);
ksynanaerobic(p) = x;
deltaR5(g,p) = diff(x);
progressbar([],p/n,4/4,[]);
end
ksynanaer(g) = mean(ksynanaerobic);
ksynanaersd(g) = sqrt((sum((ksynanaerobic-mean(ksynanaerobic)).^2))/n);
HLsd(g) = sqrt((sum((deltaR5(g,:)-mean(deltaR5(g,:))).^2))/n);
progressbar(g/lcol,0,0,0);
end
% optimalization routine
function deltaR = calcdeltaR
% indexing parameter
i=0;
for hl = hlrange
i = i+1;
% define differential equation parameters
halflife = hl;
% "
r = (1 / pulsetime)*log(growth);
% "
kdeg = log(2) / halflife;
% "
24
ksyn = r + kdeg;
options =
optimset('Display','off','Largescale','off','Algorithm','levenbergmarquardt');
%#ok<SETNU> optimize differential equations as found in the "kinetics"
function for x
[x,resnorm] = lsqnonlin(@diff,x0,lb,ub,options);
%#ok<AGROW> re-enter x in diff(x) to output(deltaR)
deltaR(i) = diff(x);
% update progress bars
if b==1
progressbar([],[],[],hl/10001);
% "
elseif b==2
[d,e] = size(hlleft:10:hlright);
progressbar([],[],1/4,i/e);
% "
elseif b==3
[d,e] = size(hlleft:hlright);
progressbar([],[],2/4,i/e);
end
end
end
% write data to Excel
disp('Writing...')
xlswrite(path, {'Half-life'}, 1, 'I1')
xlswrite(path, HL', 1, 'I2')
xlswrite(path, {'sd'}, 1, 'J1')
xlswrite(path, HLsd', 1, 'J2')
xlswrite(path, {'ksynanaer'}, 1, 'K1')
xlswrite(path, ksynanaer', 1, 'K2')
xlswrite(path, {'sd'}, 1, 'L1')
xlswrite(path, ksynanaersd', 1, 'L2')
disp('Done!')
beep
% stop and display runtime timer
Elapsedtime = toc/60;
str = ['Elapsed time: ', num2str(Elapsedtime), ' minutes'];
disp(str)
function deltaR = diff(x)
% solve the ode as described in the function 'kinetics'
options = odeset('RelTol', 1e-4);
[t,sol] = ode45(@kinetics,tspan,y0,options,x);
25
% remove y0 from data to prevent NaN errors
sol(1,:)=[];
% extract y vectors from the sol matrix
y1 = sol(:,1);
y2 = sol(:,2);
y3 = sol(:,3);
y4 = sol(:,4);
% fix
ny1 =
ny2 =
ny3 =
ny4 =
y vector lenght to prevent matrix dimension errors
y1(1:z);
y2(1:z);
y3(1:z);
y4(1:z);
% calculate the difference between measured en predicted data
Paeroob = ny1 + ny2;
Panaeroob = ny3 + ny4;
Razhal = ny4 ./ ny2;
Rtotaal = Panaeroob ./ Paeroob;
deltaR = abs(Razhal- Rnewmeasured)+abs(Rtotaal-Rtotalmeasured);
deltaR = deltaR(z);
end
%#ok<INUSL> used by ODE solver
function dydt = kinetics(t,y,x)
% define differential equations
k = [kdeg; ksyn; kdeg; x];
dydt =
[
-k(1)*y(1)
k(2)*y(1) + (k(2)-k(1))*y(2)
-k(3)*y(3)
k(4)*y(3) + (k(4)-k(3))*y(4) ];
26
Proteomic Dataset
protein no.
1
2
3
4
5
6
7
8
9
10
11
12
13
14
15
16
17
18
19
20
21
22
23
24
25
26
27
28
29
acession no.
P0A9Q7
P09373
P0AED0
P68066
P0A796
P0AET8
P37689
P0ABJ9
P0A799
P21599
P0A6T1
P0A8M6
P08839
P0A9B2
P0A6P9
P0AB71
P0A8M3
P08506
P32176
P23843
P69783
P77804
P0A707
P0A6Y8
P23893
P0ABH9
P0AD61
P17169
P0A9A6
protein name
ADHE
PFLB
USPA
GRCA
K6PF1
HDHA
GPMI
CYDA
PGK
KPYK2
G6PI
YEEX
PT1
G3P1
ENO
ALF
SYT
DACC
FDOG
OPPA
PTGA
YDGA
IF3
DNAK
GSA
CLPA
KPYK1
GLMS
FTSZ
Rnew*
10,26
7,76
6,43
6,31
5,15
4,96
4,31
3,38
3,16
3,12
3,01
2,61
2,54
2,46
2,38
2,06
2
1,93
1,93
1,84
1,81
1,77
1,75
1,66
1,63
1,61
1,61
1,57
1,52
S.D.
2,93
4,59
1,85
2,75
1,47
0,61
1,02
0,59
0,94
0,87
0,43
0,25
0,55
0,64
0,8
0,14
0,32
0,12
0,21
0,44
0,4
0,33
0,47
0,42
0,19
0,17
0,1
0,52
0,24
Rtotal**
2,59
2,28
1,5
5,45
1,81
0,95
1,41
1,72
1,34
1,22
1,21
1,37
1,07
1,21
1,14
1,12
1,07
0,94
1,32
1,14
1
0,99
0,76
1,04
1,03
1,62
1,13
1,05
1,28
S.D.
0,68
0,73
0,17
1,55
0,27
0,07
0,1
0,24
0,21
0,38
0,13
0,3
0,13
0,16
0,21
0,13
0,18
0,07
0,24
0,17
0,16
0,1
0,09
0,22
0,06
0,08
0,16
0,1
0,51
Half-life
44,3
38,9
203,1
3
39,4
10000
108
20,8
73,2
287,9
301,6
36,6
10000
118,4
3564,2
10000
10000
10000
20,2
139,1
10000
10000
10000
10000
10000
1,2
88,3
10000
9,9
S.D.
0,51
0,66
0,13
1,61
0,22
0,08
0,09
0,17
0,15
0,22
0,07
0,19
0,08
0,09
0,13
0,07
0,11
0,07
0,15
0,10
0,11
0,08
0,09
0,13
0,05
0,11
0,10
0,07
0,33
ksynanaer
0,123
0,103
0,046
0,433
0,085
0,026
0,044
0,096
0,040
0,023
0,022
0,053
0,014
0,025
0,013
0,012
0,011
0,011
0,065
0,018
0,010
0,010
0,010
0,010
0,009
0,650
0,020
0,009
0,098
S.D.
0,025
0,061
0,012
0,134
0,019
0,003
0,009
0,012
0,010
0,006
0,003
0,004
0,003
0,006
0,004
0,001
0,002
0,001
0,005
0,004
0,002
0,002
0,002
0,002
0,001
0,016
0,001
0,003
0,010
protein no.
30
31
32
33
34
35
36
37
38
39
40
41
42
43
44
45
46
47
48
49
50
51
52
53
54
55
56
57
58
59
60
61
62
63
acession no.
P0AFH8
P0A6H1
P0ABT2
P09169
P0A9X4
P23836
P0A817
P0A905
P0A910
P0C0S1
P06959
P0AEP3
P00509
P0A953
P61714
P0A870
P25665
P69776
P0A825
P0ACF8
P00956
P00562
P0AFG8
P00350
P0ABB4
P0A9P0
P0AEG4
P0AEK4
P0AES4
P0A705
P07813
P23721
P0A8M0
P68919
protein name
OSMY
CLPX
DPS
OMPT
MREB
PHOP
METK
SLYB
OMPA
MSCS
ODP2
GALU
AAT
FABB
RISB
TALB
METE
LPP
GLYA
HNS
SYI
AK2H
ODP1
6PGD
ATPB
DLDH
DSBA
FABI
GYRA
IF2
SYL
SERC
SYN
RL25
Rnew*
1,46
1,42
1,4
1,35
1,3
1,3
1,26
1,12
1,11
1,07
1,04
1,04
1,01
0,97
0,96
0,95
0,95
0,95
0,91
0,9
0,87
0,86
0,84
0,82
0,77
0,77
0,77
0,76
0,76
0,74
0,73
0,72
0,71
0,66
S.D.
0,24
0,37
0,29
0,35
0,47
0,21
0,37
0,23
0,36
0,15
0,39
0,11
0,12
0,26
0,15
0,11
0,3
0,14
0,41
0,21
0,13
0,17
0,23
0,1
0,14
0,12
0,11
0,11
0,07
0,1
0,4
0,1
0,16
0,37
Rtotal**
1,07
0,93
1,38
0,91
0,92
1,03
0,88
0,95
0,99
0,99
1
1,05
1,02
0,99
0,94
0,93
0,6
0,79
1,05
1,08
0,84
0,75
0,98
0,98
1,02
0,97
1,1
0,96
0,93
0,91
1,01
1,12
0,88
0,98
S.D.
0,12
0,15
0,22
0,26
0,14
0,21
0,04
0,13
0,15
0,1
0,14
0,07
0,12
0,07
0,08
0,13
0,04
0,13
0,16
0,26
0,15
0,02
0,16
0,17
0,15
0,16
0,27
0,07
0,1
0,16
0,23
0,23
0,15
0,13
Half-life
10000
10000
1,9
9982,3
10000
10000
10000
9981,2
9930,9
9940,9
1,2
7,8
3,5
3,9
1,2
1,2
1,2
1,2
278,4
10000
1,2
1,2
15,2
17,4
67,4
19,7
10000
17,6
11,6
10,2
58,1
10000
8,8
37,8
S.D.
0,07
0,13
0,21
0,17
0,11
0,12
0,04
0,10
0,11
0,07
0,23
0,06
0,10
0,14
0,10
0,10
0,25
0,15
0,10
0,15
0,11
0,11
0,12
0,11
0,08
0,10
0,15
0,05
0,06
0,10
0,15
0,15
0,11
0,10
ksynanaer
0,008
0,008
0,399
0,008
0,008
0,007
0,007
0,006
0,006
0,006
0,567
0,094
0,198
0,173
0,566
0,565
0,563
0,567
0,007
0,005
0,551
0,545
0,041
0,037
0,012
0,031
0,005
0,034
0,051
0,056
0,012
0,004
0,062
0,016
S.D.
0,001
0,002
0,028
0,002
0,003
0,001
0,002
0,001
0,002
0,001
0,074
0,007
0,012
0,027
0,024
0,018
0,059
0,025
0,003
0,001
0,023
0,033
0,010
0,004
0,002
0,004
0,001
0,004
0,004
0,006
0,007
0,001
0,011
0,009
protein no.
64
65
66
67
68
69
70
71
72
73
74
75
76
77
78
79
80
81
82
83
84
85
86
87
88
89
90
91
92
93
94
95
96
97
acession no.
P0ABB0
P0AFG0
P0AEU7
P13029
P0AE08
P0A8T7
P0A6N1
P30750
P0A6P1
P0AG30
P0ABJ1
P45577
P0A850
P0A7D7
P0A6M8
P0A7V0
P0A8V2
P0AG48
P0A7W1
P05055
P0A6Q3
P62399
P35340
P0AG44
P60438
P0AG67
P0A7X3
P0A7R1
P0AA10
P0AG55
P0A912
P0A7L0
P0A7U7
P02359
protein name
ATPA
NUSG
SKP
KATG
AHPC
RPOC
EFTU
METN
EFTS
RHO
CYOA
PROQ
TIG
PUR7
EFG
RS2
RPOB
RL21
RS5
PNP
FABA
RL5
AHPF
RL17
RL3
RS1
RS9
RL9
RL13
RL6
PAL
RL1
RS20
RS7
Rnew*
0,66
0,66
0,66
0,64
0,64
0,61
0,59
0,58
0,58
0,58
0,55
0,55
0,53
0,53
0,5
0,47
0,45
0,44
0,43
0,43
0,42
0,39
0,37
0,36
0,35
0,35
0,34
0,34
0,32
0,31
0,31
0,31
0,3
0,3
S.D.
0,1
0,04
0,12
0,19
0,46
0,19
0,1
0,07
0,09
0,12
0,05
0,04
0,16
0,09
0,11
0,04
0,07
0,04
0,2
0,11
0,06
0,15
0,16
0,07
0,06
0,05
0,07
0,03
0,06
0,06
0,06
0,09
0,04
0,01
Rtotal**
0,99
0,93
0,95
0,67
0,62
0,97
0,94
0,62
0,94
0,97
1,16
0,81
0,94
0,94
0,97
0,93
0,96
0,9
0,9
0,75
0,93
0,9
0,49
0,83
0,85
1,02
0,92
0,93
0,95
0,94
0,74
0,89
0,83
0,98
S.D.
0,17
0,02
0,08
0,09
0,2
0,19
0,15
0,03
0,11
0,07
0,29
0,08
0,12
0,18
0,15
0,13
0,15
0,26
0,12
0,17
0,1
0,11
0,14
0,11
0,15
0,2
0,18
0,16
0,11
0,08
0,13
0,15
0,17
0,17
Half-life
46,5
18,4
23,7
2,1
1,2
38
26,5
2,2
27,3
42
10000
9,6
31,7
31,7
54,2
33,1
52,6
26,2
26,8
9,5
37,5
29,4
3,3
18,2
21,5
7898,4
40,2
45,2
62,5
55,1
12
31,8
20,8
124,7
S.D.
0,10
0,01
0,05
0,12
0,26
0,12
0,10
0,04
0,06
0,04
0,18
0,05
0,08
0,11
0,08
0,07
0,09
0,15
0,08
0,11
0,06
0,07
0,10
0,07
0,08
0,11
0,11
0,09
0,06
0,05
0,07
0,08
0,10
0,10
ksynanaer
0,013
0,029
0,023
0,268
0,406
0,014
0,019
0,250
0,018
0,013
0,003
0,047
0,015
0,015
0,009
0,013
0,009
0,015
0,014
0,038
0,010
0,012
0,115
0,017
0,014
0,002
0,008
0,007
0,005
0,006
0,023
0,009
0,013
0,003
S.D.
0,002
0,002
0,004
0,037
0,309
0,004
0,003
0,014
0,002
0,003
0,000
0,003
0,004
0,002
0,002
0,001
0,001
0,001
0,006
0,009
0,001
0,004
0,034
0,003
0,002
0,000
0,002
0,001
0,001
0,001
0,004
0,002
0,002
0,000
protein no.
acession no. protein name
Rnew*
S.D.
Rtotal**
S.D.
Half-life
S.D.
ksynanaer
98
P0ABK5
CYSK
0,29
0,02
0,51
0,08
4,8
0,05
0,062
99
P60422
RL2
0,28
0,1
0,86
0,13
26,5
0,08
0,010
100
P0A7R5
RS10
0,26
0,07
0,92
0,17
47,6
0,11
0,005
101
P0A7W7
RS8
0,26
0,01
0,91
0,07
42,6
0,05
0,006
102
P60723
RL4
0,26
0,05
0,86
0,13
27,6
0,08
0,008
103
P0A7J7
RL11
0,21
0,06
0,95
0,27
81
0,16
0,003
* ratio of newly synthesized proteins made during the pulse with azhal, ** ratio of protein level changes following a switch to an anaerobic environment
S.D.
0,003
0,003
0,001
0,000
0,002
0,001
References
1
Lodish, H., Berk, A., Matsudaira, P., Kaiser, C.A., Krieger, M., Scott, M.P., Zipurksy, S.L.,
Darnell, J. , (2004) Molecular Cell Biology
2
Branden, C. and Tooze, J., (1999) Introduction to Protein Structure
3
Murray et al., (2009) Harper's Illustrated Biochemistry
4
Stepanenko, O.V., Verkhusha , V.V., Kuznetsova, I.M., Uversky, V.N., Turoverov, K.K.
(2008)Fluorescent proteins as biomarkers and biosensors: throwing color lights on
molecular and cellular processes
5
Margolin, W. (2000) Green fluorescent protein as a reporter for macromolecular
localization in bacterial cells
6
Görg, A., Weiss, W., Dunn, M.J. (2004) Current two-dimensional electrophoresis
technology for proteomics
7
Conrotto, P., Souchelnytskyi, S. (2008) Proteomic approaches in biological and medical
sciences: principles and applications
8
http://en.wikipedia.org/wiki/Computational_systems_biology, 19/12/2011 14:33
9
Koster, C., Kramer, G., Koning, L., Jong, L., (2010) A mathematical model for the
estimation of protein half-lifes in pulse labeling proteomics experiments.
10
Kramer, G., Sprenger, R., Nessen, M., Roseboom, W., Speijer, D., Jong, L., Teixeira de
Mattos, J., Back, J.,W., Koster, C. (2010) Proteome-wide alterations in Escherichia coli
translation rates upon anaerobiosis.
11
Kramer, G., Sprenger, R., Nessen, M., Jong, L., Back, J., W., Koster, C., Dekker, H.,
Maarseveen, J., Koning, L., Hellingwerf, K., (2009) Identification and Quantitation of Newly
Synthesized Proteins in Escherichia coli by Enrichment of Azidohomoalanine-labeled
Peptides with Diagonal Chromatography.
12
Larrabee K. L., Phillips J. O.,Williams G. J., Larrabee A. R. (1980) The relative rates of
protein synthesis and degradation in growing culture of Escherichia coli
13
Mostellerr, D., Goldstein, R. V., Nishimoto, K. R. (1980). Metabolism of individual
proteins in exponentially growing Escherichia coli.
31