Crash-Kurs Molekulare Evolution
advertisement
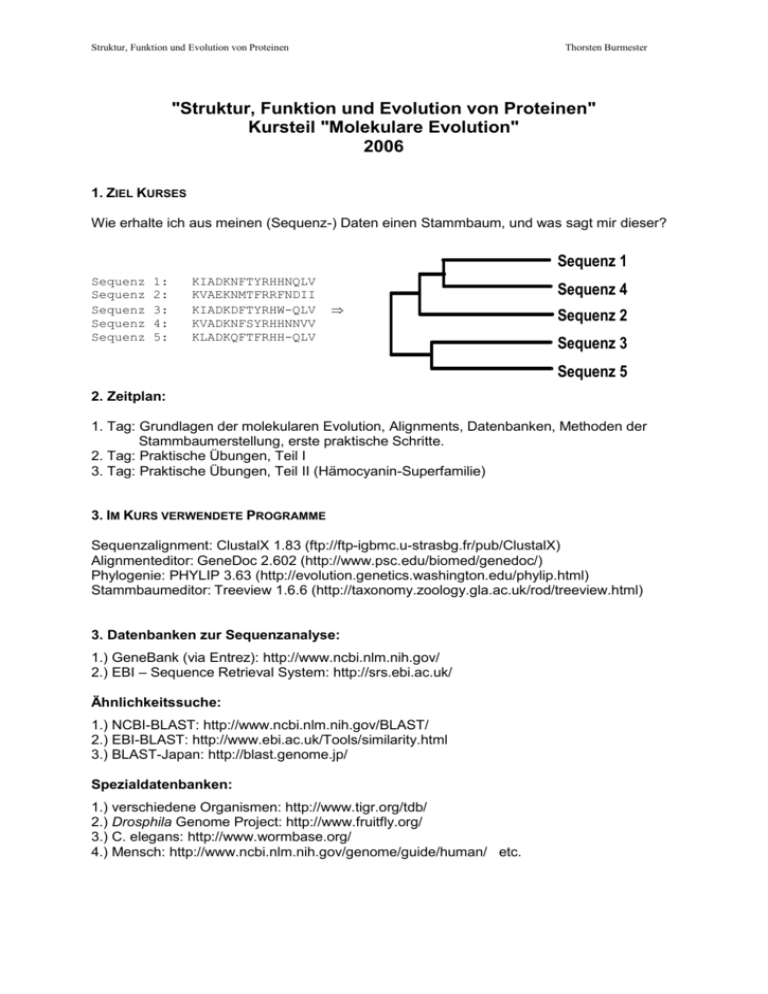
Struktur, Funktion und Evolution von Proteinen Thorsten Burmester "Struktur, Funktion und Evolution von Proteinen" Kursteil "Molekulare Evolution" 2006 1. ZIEL KURSES Wie erhalte ich aus meinen (Sequenz-) Daten einen Stammbaum, und was sagt mir dieser? Sequenz 1 Sequenz Sequenz Sequenz Sequenz Sequenz 1: 2: 3: 4: 5: KIADKNFTYRHHNQLV KVAEKNMTFRRFNDII KIADKDFTYRHW-QLV KVADKNFSYRHHNNVV KLADKQFTFRHH-QLV Sequenz 4 Sequenz 2 Sequenz 3 Sequenz 5 2. Zeitplan: 1. Tag: Grundlagen der molekularen Evolution, Alignments, Datenbanken, Methoden der Stammbaumerstellung, erste praktische Schritte. 2. Tag: Praktische Übungen, Teil I 3. Tag: Praktische Übungen, Teil II (Hämocyanin-Superfamilie) 3. IM KURS VERWENDETE PROGRAMME Sequenzalignment: ClustalX 1.83 (ftp://ftp-igbmc.u-strasbg.fr/pub/ClustalX) Alignmenteditor: GeneDoc 2.602 (http://www.psc.edu/biomed/genedoc/) Phylogenie: PHYLIP 3.63 (http://evolution.genetics.washington.edu/phylip.html) Stammbaumeditor: Treeview 1.6.6 (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html) 3. Datenbanken zur Sequenzanalyse: 1.) GeneBank (via Entrez): http://www.ncbi.nlm.nih.gov/ 2.) EBI – Sequence Retrieval System: http://srs.ebi.ac.uk/ Ähnlichkeitssuche: 1.) NCBI-BLAST: http://www.ncbi.nlm.nih.gov/BLAST/ 2.) EBI-BLAST: http://www.ebi.ac.uk/Tools/similarity.html 3.) BLAST-Japan: http://blast.genome.jp/ Spezialdatenbanken: 1.) verschiedene Organismen: http://www.tigr.org/tdb/ 2.) Drosphila Genome Project: http://www.fruitfly.org/ 3.) C. elegans: http://www.wormbase.org/ 4.) Mensch: http://www.ncbi.nlm.nih.gov/genome/guide/human/ etc. Struktur, Funktion und Evolution von Proteinen Thorsten Burmester ANHANG: 1. PROGRAMME ZUR MOLEKULAREN PHYLOGENIE: (im Folgenden werden, bis auf eine Ausnahme, nur DOS oder Windows-Programme berücksichtigt. Diese Liste ist selbstverständlich sehr unvollständig und erwähnt nur die am häufigsten gebrauchten Programme. Siehe auch: http://www.ebi.ac.uk/biocat/ (veraltet!), oder http://evolution.genetics.washington.edu/phylip/software.html (aktuell). z.B.: 1.1 ALIGNMENT VON ZWEI SEQUENZEN: FASTA: ftp://ftp.bio.indiana.edu/molbio/search/ ALIGN: http://www2.igh.cnrs.fr/bin/align-guess.cgi LALIGN: http://www2.igh.cnrs.fr/bin/lalign-guess.cgi 2.2 MULTIPLE SEQUENCE ALIGNMENT: ClustalX, aktuelle Version 1.83: ftp://ftp-igbmc.u-strasbg.fr/pub/ClustalX PILEUP (GCG-Sofware-Package): UNIX-Program, man benötigt Zugang zum gesamten GCG-Packet. 2.3 MULTIPLE SEQUENCE ALIGNMENTS EDITOR: GeneDoc, aktuelle Version 2.602: http://www.psc.edu/biomed/genedoc/ Sehr gutes Programm, kann alles, was ein MSF-Editor können muß. Liest alle üblichen MSF-Formate, erlaubt Editieren, Sekundärstrukturanalyse. 2.4 PROGRAMME ZUR STAMMBAUMERSTELLUNG: ClustalX (s.o.): erlaubt die Erstellung einfacher NJ Stammbäume PHYLIP, aktuelle Version 3.6b: http://evolution.genetics.washington.edu/phylip.html Programmpacket, besteht aus vielen Einzelprogrammen zur Analyse eine Aminosäure oder DNA MSF. Matrixanalyse, Distanzberechnungen; NJ, Least-Squares Methode, MP, ML PAUP, aktuelle Version 4: kommerzielles Programm, zur Zeit nur als beta-Version erhältlich soll irgendwann bei Sinauer erscheinen. Erstellt MP, ML und NJ Stammbäume Tree-Puzzle, aktuelle Version 5.1: http://www.tree-puzzle.de/ ML-Programm; quartet puzzling; MOLPHY aktuelle Version 2.2: http://dogwood.botany.uga.edu/malmberg/software.html ML-Programm MrBayes: http://morphbank.ebc.uu.se/mrbayes/ Bayes'sche Phylogenie 2.5 STAMMBAUMEDITOREN Treeview, aktuelle Version 1.6.6: http://taxonomy.zoology.gla.ac.uk/rod/treeview.html NJplotWIN95: ftp://biom3.univ-lyon1.fr/pub/mol_phylogeny/njplot TreeCon: http://www.evolutionsbiologie.uni-konstanz.de/peer-lab/treeconw.html TreeExplorer: http://evolgen.biol.metro-u.ac.jp/TE/TE_man.html Struktur, Funktion und Evolution von Proteinen Thorsten Burmester 2. LITERATUR: 2.1 Bücher: W.-H. Li and D. Graur, Fundamentals of Molecular Evolution, Sinauer; 1991. W.-H. Li, Molecular Evolution, Sinauer, 1997. D. M. Hillis, C. Moritz, and Mabel (Eds.), Molecular Systematics, Sinauer; 1996 Page, R. D. M and Holms, Molecular Evolution: A Phylogenetic Approach, 1998, Blackwell Science Wägele, J.-W.: Grundlagen der Phylogenetischen Systematik, Pfeil-Verlag, 2001 Felsenstein, J., Inferring Phylogenies, Sinauer, 2004 Hall, B. G. Phylogenetic Trees Made Easy, Sinauer, 2004 2.2 WWW: http://www.lmb.uni-muenchen.de/groups/bioinformatics/bioinfo.html http://www.hgmp.mrc.ac.uk/MANUAL/faq/faq-phylogeny.html ... und vieles mehr! Struktur, Funktion und Evolution von Proteinen Thorsten Burmester 3. GLOSSAR: Introductory Glossary of Cladistic Terms von: Michael D. Crisp Division of Botany and Zoology Australian National University Canberra ACT 2601 http://www.science.uts.edu.au/sasb/glossary.html Additive For application to characters, see ordered. As applied to trees, it refers to whether distances measured along the branches of the tree add up to the observed distances (from a matrix of pair-wise distance comparisons among terminal taxa). Alignment The determination of positional homology for molecular sequences, involving the juxtaposition of amino acids or nucleotides in homologous molecules. Anagenesis See phylesis. Analogy See homoplasy. Apomorphy (adj. apomorphic or apomorphous) A relatively derived or advanced or unique character state (cf. autapomorphy, synapomorphy, plesiomorphy, symplesiomorphy). Area cladogram A tree that displays historical relationships among geographic areas, rather than phylogenetic relationships among taxa. Attribute The possession by an organism of a particular feature, e.g. this tree is rough-barked, that tree is half-barked (cf. character and character states). Autapomorphy An apomorphy in a terminal taxon; diagnoses the terminal but is uninformative about relationships to other terminals; therefore of no use for cladistic tree-building. Binary A character type with only two states (usually given as 0, 1), in which a change in either direction between the states is 1 step (cf. ordered, unordered, Dollo, irreversible). Character Any heritable attribute of organisms that varies among terminal taxa, and so is useful for phylogenetic reconstruction. Character states Subdivisions of the variation among terminal taxa. Clade A monophyletic group (= a branch on a cladogram, diagnosed by at least one synapomorphy). Cladogenesis The evolutionary splitting of lineages, i.e. speciation (cf. phylesis). Cladogram A branching diagram (tree) assumed to be an estimate of a phylogeny (cf. phylogram, dendrogram, phenogram). Classification Arranging organisms into named groups (taxa), whether natural or artificial (see systematisation). Congruence (adj. congruent) Agreement, as between characters and a tree, or between the topologies (shapes) of two trees, e.g. derived from different data sets, such as molecular and morphological. Struktur, Funktion und Evolution von Proteinen Thorsten Burmester Some authors like to make separate phylogeny estimates from different data sets, and then test their congruence (cf. total evidence). Consensus A class of methods used to estimate the amount of agreement among incongruent or partially congruent trees. Usually represented as a tree that is less resolved than any of the input trees. (There are also consensus statistics.) A consensus tree is not an hypothesis of evolutionary history, and thus must not be confused with a phylogenetic tree. Therefore, it should not be used to trace evolution of characters, areas (biogeography), and so on. Most commonly used is the strict consensus tree, which shows only those clades that are common to all the input trees; a majority-rule consensus tree shows all clades that are found in > 50% of the input trees. Consistency index (CI) A measure of the parsimony fit of a character to a tree, or of the average fit of all characters to a tree. Varies from 1.0 (perfect fit) to a value asymptotically approaching zero (poorest fit). It is inflated by autapomorphies which can only take the value 1.0; thus a totally uninformative data set (consisting only of autapomorphies) could return a CI equal to 1.0 (cf. retention index). Convergence See homoplasy. Daughter taxa See sister groups. Dendrogram Any branching diagram (or tree) (cf. cladogram, phylogram, phenogram). Distance Usually treated as a measure of evolutionary divergence, i.e. phylogenetic distance increases with increasing evolutionary divergence. Distances are usually expressed pair-wise among the terminal taxa, and can be calculated based on a specified evolutionary model; the model specifies the probabilities of character-state changes through evolutionary time. Distances are popular for building phylogenetic trees from molecular sequence data (cf. maximum likelihood, parsimony). Dollo A character type in which numerically increasing changes are allowed but each such change can only happen once on a tree; thus, multiple reverse changes (= losses) are allowed. This character type is favoured by those who feel that a complex structure (e.g. the insect wing) can only originate once, although it may be lost many times. This character type has been suggested for DNA restriction site data, because gain of a new site is much more improbable than loss of an existing one. By definition, a Dollo character is polarised in advance, making the use of an outgroup redundant (cf. ordered, unordered, irreversible). Exact method Any analysis method that guarantees to find the optimal solution. For tree-building, the branch-and-bound strategy is a computationally-efficient exact method for finding the optimal tree that does not involve examining every possible tree (cf. heuristic method). Gene tree A phylogeny of a gene, which may or may not accurately reflect the phylogeny of the organisms possessing that gene (see orthology). Heuristic method Any analysis method involving computationally-efficient strategies that should produce a solution at least close to the optimal one even if it doesn't find the optimum (cf. exact method). Homology (adj. homologous) Similarity due to common evolutionary origin, i.e. derived from the same ancestral character; thus, equivalent to synapomorphy. Morphologists also define homology by common developmental origin, which is quite a different concept, being based on Struktur, Funktion und Evolution von Proteinen Thorsten Burmester a different process, although empirically the two homologies may be congruent. Noncladists like to include symplesiomorphy in their concept of homology. Homoplasy (adj. homoplastic or homoplasious) Similarity due to independent evolutionary change. Thus, homoplasy is a mistaken hypothesis of homology, which will confound cladistic analyses. Homoplasy is either parallelism (= independent gain) or reversal (= loss). Convergence (= analogy) is sometimes distinguished from parallelism, although the distinction may be arbitrary (and in practice the difference may be irrelevant). Convergent features are derived from distantly-related ancestors, e.g. the wings of bats and birds, or succulence in Cactaceae and Euphorbiaceae (i.e. independent evolution derived by a different mechanism, thus leading to superficial similarity). Parallelisms derive from closelyrelated ancestors, e.g. the nucleotide A derived independently in two descendant lineages from the same C in the same position in a DNA sequence in a common ancestor (i.e. independent evolution using the same mechanism). Convergent features can usually be distinguished by detailed examination (e.g. differences in internal anatomy), whereas in the nucleotide example this would be impossible. Informative Refers to the part of the data that is actually used by a particular method for building trees (cf. uninformative). Ingroup The study group whose phylogeny is being reconstructed (cf. outgroup). Irreversible (Camin-Sokal) A character type in which numerically increasing changes are allowed and counted as for ordered characters, while decreasing changes are not allowed (i.e. counted as an infinite number of steps); thus, multiple reverse changes (= losses) are not allowed. By definition, an irreversible character is polarised in advance, making the use of an outgroup redundant. This character-type is very rarely used, as the assumption of irreversibility is very difficult to justify for any type of data, morphological or molecular. It was proposed by E.O. Wilson (1965, Systematic Zoology 14:214-220), with examples of its application (cf. ordered, unordered, Dollo). Lineage An historical sequence of ancestors and descendants. Maximum likelihood One of several criteria that may be optimised in building phylogenetic trees from molecular sequence data. The optimal tree is the one that maximises the statistical likelihood that the specified evolutionary model produced the observed characterstate data; the models specify the probabilities of character-state changes through evolutionary time (cf. distance, parsimony). Monophyly (holophyly) (adj. monophyletic, holophyletic) On a phylogeny, a monophyletic group has a unique origin in a single ancestral species, and includes the ancestor and all of its descendants. It is recognised by a homologous character state (synapomorphy) in all of its members (cf. paraphyly, polyphyly). Network See unrooted tree. Node A branch-point on a tree / cladogram. Non-additive See unordered. Ordered (additive) A character type with > 2 states that follow an evolutionarily plausible sequence, e.g. petals many -> 5 -> 3 -> 0. Changes between adjacent states are counted as one step and changes between non-adjacent states are counted as (1 + no. of skipped states), e.g. from 5 petals to 0 (or vice versa) would be 2 steps (cf. unordered, Dollo, irreversible). Orthology Struktur, Funktion und Evolution von Proteinen Thorsten Burmester True homology of molecular sequences, i.e. descended in toto from the same ancestral sequence. Orthologous sequences exist in only one copy per organism, and can accurately reflect the phylogenetic relationships of species (cf. paralogy, plerology, xenology). Outgroup A terminal taxon (or group of taxa), preferably the sister-group of the ingroup, that is used to root a cladogram (cf. ingroup). The root is placed between the outgroup(s) and the ingroup. Multiple outgroups may be used. Parallelism See homoplasy. Paralogy Paralogous molecular sequences result from gene duplication (independent of organism speciation), exist in multiple copies per organism, and will reconstruct gene phylogeny rather than species phylogeny (which may not be congruent) (cf. orthology). Paraphyly (adj. paraphyletic) A paraphyletic group originates from a single common ancestor, which is included in the group, but does not include all of the descendants of that ancestor (cf. monophyly, polyphyly). Its members share only ancestral character states (symplesiomorphies); they do not uniquely share any synapomorphies. Parsimony One of several criteria that may be optimised in building phylogenetic trees, but a philosophically important one due to its simplicity; and the basis of the mostcommonly used method of cladistic analysis, at least for morphological data. The central idea of cladistic parsimony analysis is that some trees will fit the characterstate data better than other trees. Fit is measured by the number of evolutionary character-state changes implied by the tree. The fewer changes the better, e.g. there is no sense in choosing a phylogeny that has roots, flowers and xylem each evolving twice, if another tree exists on which one evolutionary origin for each of the apomorphic states would explain the observed distribution of states across taxa(cf. distance, maximum likelihood). Phenetic Similarity of characters without regard to the distinction between synapomorphy, homoplasy and symplesiomorphy. Phenetic methods are poor at reconstructing phylogeny. Phenogram A branching diagram (tree) showing the phenetic similarity among the terminal taxa (cf. cladogram, phylogram, dendrogram). Phylesis (anagenesis) Evolutionary events that modify a taxon without causing speciation (cf. cladogenesis). Phylogeny The unique historical relationship (resulting from evolution) among terminal taxa, represented as a tree (cf. cladogram). Phylogram A branching diagram (tree) assumed to be an estimate of a phylogeny; usually distinguished from a cladogram in that the branch lengths are proportional to the amount of inferred evolutionary change (cf. cladogram, phenogram, dendrogram). Plerology Partial homology of molecular sequences resulting from an inter-mixture of exons and introns; will only reconstruct a composite gene history (cf. orthology). Plesiomorphy A relatively primitive or ancestral character state (cf. apomorphy). Polarity Struktur, Funktion und Evolution von Proteinen Thorsten Burmester Evolutionary ordering of character states, determined either independently of tree construction (direct method) or more usually from a rooted phylogenetic tree (indirect method). Polyphyly (adj. polyphyletic) A polyphyletic group does not include a unique common ancestor, i.e. it has multiple evolutionary origins. This concept is best restricted to groups of hybrid origin, e.g. eukaryotes, allopolyploids; otherwise, the distinction from paraphyly is somewhat arbitrary, since inclusion / exclusion of the ancestor would be the only difference (cf. monophyly, paraphyly). Polytomy (polychotomy) A branch-point in a tree with more than two descendant branches. A polytomy referred to as "hard" results from absence of data to resolve branching dichotomously, and may be interpreted as multiple speciation. A polytomy referred to as "soft" reflects uncertainty resulting from conflict (incongruence) among two or more fully-resolved cladograms. Retention index (RI) Similar to the consistency index, but defined so that the highest possible value for any character is 1.0 and the lowest is 0.0; removes bias due to autapomorphies (cf. consistency index). Reversal (= loss) Evolutionary reversion from an apomorphic to a plesiomorphic character state (cf. homoplasy). Rooted tree A cladogram with a hypothetical ancestor, which equates to the root, which is the node at the base of the tree. When outgroups are used, this is the node that connects the outgroups to the ingroup, and which thus specifies the direction of evolutionary change among the character-states (cf. unrooted tree). Sister groups (or taxa) The descendant branches from a node on a cladogram. In a phylogeny, the descendants of an ancestor are called daughters, while the siblings after a speciation event are called sisters (so a descendant is a daughter relative to its ancestor and is a sister relative to its other sibling). Note that if either of the daughters undergoes further speciation then the sister to a particular terminal taxon may actually be a group of terminal taxa. Symplesiomorphy A plesiomorphy shared by two or more terminal taxa, only diagnostic of a paraphyletic group (cf. synapomorphy). Synapomorphy An apomorphy shared by two or more terminal taxa; thus diagnoses a clade or monophyletic group (see also homology). Speciation The evolutionary splitting of lineages. Species Difficult to define rigorously in two or three lines. Defined very simply in a phylogenetic context, species are the smallest lineages that are mutually exclusive of other lineages. The internal branches of a phylogeny may be viewed as ancestral species. Note, however, that the unit lineages of a gene phylogeny are not species (see also terminal). Step A single character-state change. Systematisation Reconstructing natural (i.e. phylogenetic) relationships among organisms (cf. classification). Taxon (pl. taxa) A named group of organisms, not necessarily a natural (monophyletic) unit (cf. terminal). Struktur, Funktion und Evolution von Proteinen Thorsten Burmester Terminal (terminal taxon) One of the units whose collective phylogeny is reconstructed; in other words, the undivided tips of a tree (usually contemporary taxa). Terminals may be higher taxa, species, populations, individuals, fossils or even genes. There should be some rational basis for accepting the integrity of each terminal (for the purpose of the analysis), e.g. a monophyletic or diagnosable unit. Despite the claims by some authors, terminals do not need to be monophyletic; in fact, many species-level terminals are unavoidably paraphyletic. However, higher taxa used as terminals should be monophyletic. Topology The branching sequence of a tree. Total evidence Reconstructing phylogeny by analysing combined data of different kinds, e.g. morphology and gene sequences. A controversial issue, because gene phylogenies may be incongruent with organismal phylogenies (cf. congruence). Tree Mathematically, an acyclic (cycle-free) line graph. Used to represent the evolutionary history of a set of taxa, with the leaves (or terminal branches) representing contemporary taxa and the internal branches representing hypothesised ancestors (see also rooted tree, unrooted tree). Uninformative All tree-building methods discard some data, and therefore such data are "uninformative" for building trees using that method. For instance, in parsimony methods only characters whose number of steps can vary on trees are informative; autapomorphic and invariant characters are uninformative (these can be determined by inspection of the data). However, in UPGMA autapomorphic characters are informative. (cf. informative). Unordered (non-additive) A character type with > 2 states that have no plausible evolutionary sequence, e.g. the nucleotides A, C, G and T. A change between any pair of states is counted as 1 step. This is by far the most common type of character state used in cladistic analyses (cf. ordered, Dollo, irreversible). Unrooted tree (network) A cladogram for which the ancestor (= root) has not been hypothesized, and which thus does not specify the direction of evolutionary change among the characterstates. An unrooted tree can be rooted on any of its branches, and so there are many rooted trees that can be derived from a single unrooted tree (cf. rooted tree). Xenology A polyphyletic relationship among molecular sequences resulting from horizontal gene transfer (cf. orthology).