JCB_21826_sm_suppmat
advertisement

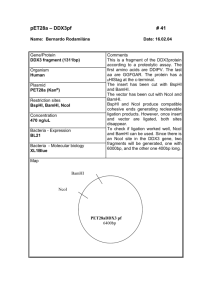
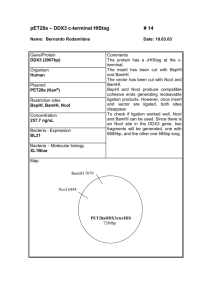
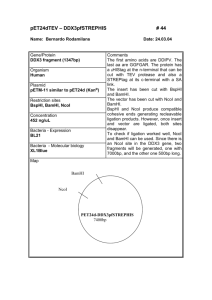
Supplementary data I. Supplementary Figs Supplementary Fig1 Confirming the digestion efficiency of cross-linked DNA in 3C assay by Agarose electrophoresis, southernblotting, semi-quantitative PCR. In the electrophoresis experiment, freshly prepared mouse genome DNA was the control. CL DNA/NcoI represent the DNA extracted from the cross linked and NcoI digested nuclei of mouse fetal liver cells. The following probes were used in southernblotting: HS8, a 502-bp PCR fragment, which hybridizes to a 2.2-kb NcoI HS8 fragment, a 485-bp PCR fragment, which hybridizes to a 2.2-kb NcoI fragment; 1, a 388-bp PCR fragment, which hybridizes to a 2.2-kb NcoI 1 fragment; 2, a 420-bp PCR fragment, which hybridizes to a 1.5-kb NcoI 2 fragment. The primers for amplifying the probes are as follows: HS8L, GATCTACAGACTGCCCTCCCAAGTC; HS8-R, TATAAAGTGCTTTCCCTCACCAGGG; -L, CATAGCCATTTGTTGCCAATCAGTG; -R, GGGCTTCATAGTGAGACCGCATC; 1-L, TGCTCACATCCATTCAGACACAGAC; 1-R, AGGTTGGGACAAGTACAGTTAGGG; 2-L, GCTGCCCTTCCCTCATCCTCTG; 2-R, AAATCCGGTTG TTACTTGATCATGC. In the semiquantitative PCR assay, the primers were designed at the two sides of NcoI sites. The mouse genome DNA is the positive control and the mouse genome completely digested by NcoI is the negative control. 1 Supplementary Fig2 Position of 475A8 on mouse chromosome 11. The position of captured 475A8 RCF belongs to BAC clone RP23-475A8. The whole BAC clone includes more than 200kb DNA sequence. There is one functional gene Fam44B in the whole BAC clone which is essential for the mitotic spindles assembly. The numbers corresponding to the sequence number of NCBI Mapviewer number. The 475A8 RCF is nearly 1.7kb and the Vista program identified two closely located highly conserved regions, CR1 and CR2, were located at the position from 0.7kb to 1.3kb of this RCF. Supplementary Fig3 Confirmation of GATA-1 expression of GATA-1 expression vector in GP293 cells. GP293 is a cell line that has no GATA-1 expression. RT-PCR was performed to detect 2 the GATA-1 expression after the GP293 cells being transfected by GATA-1 expression vector. The total RNAs were the negative control. Supplementary Fig4 Increased -globin gene expression in MEL cells induced by DMSO. RTPCR was performed to confirm that -globin gene expression increased accompany with the DMSO BAC NAME RP24-163H9 RP24-367P22 RP23-107I8 RP23-188H3 RP23203H20 RP23398G11 RP23386K17 TAG POSITION -75675 113380 22729 6935 55049 CHROMOSOME TAG SEQUENCE 15 15 10 11 10 CCATGGTTCAAAGGAAGAGAAA CCATGGACTCTCCATTTATCT CCATGGCTTTCTGTTATATG CCATGGCACCCAAGAGGTCCT CCATGGCTTTGTCACCAGGTT 3C SIGNAL/42CYCLES yes yes yes yes yes 148314 11 CCATGGCTTTGCTTTGCTTGGATTC yes -52307 11 CCATGGTTTTTGCATCCGCTCCTGCCTT yes induction of MEL cells for 1 day to 4 days. GAPDH was the control. 3 RP23-12D16 RP24175C20 RP23-30H7 113165 -136021 8 9 211765 14 CCATGGAGGCAGATCACATGCT CCATGGAAAGGTGTTTTTGC yes no CCATGGAGCCATCCGTTCTCT no II Supplementary Table Supple table1 3C verification of the low frequency tags. The 3C assay of 10 randomly selected low frequency tags. The PCR is performed as following: one cycle at 94°C for 4 min; 42 cycles at 94°C for 30 s, 58°C for 40 s and 72°C for 30 s; followed by one cycle at 72°C for 10 min. Position number represents the tag position in the BAC sequence. Tag sequences are all beginning with NcoI recognition sequences that represent the original NcoI digestion site during 3C template preparation. III. Primers used in 3C assay Primer sequences for the tested restriction fragments were as follows: HS26, GTATGGTAGCTTACCCCTATAATGCC ;475A8, AGGAGAAAGGTCACCCATGTCA ; MPG, CACAGAA -globin, -globin, GACAGTGCAGGTCTGGATACAAGA. Errc3-1 (CTATATTCAACTGCTGTTCCCATG) and Errc3-2 (TTCTACCAGCAGATC CGTATTCC) IV. Step by step description of QACT Step 1, Crosslink the cells. Purpose, fix the in vivo chromatin status. Step2, NcoI, the first enzyme digestion and religation Purpose, generating the cohesive ends by NcoI that could religate to form the hybrid molecules (spatially proximal DNA will join together) Step3, Reverse Crosslink Purpose, to purify the hybrid molecules for next step including either 3C PCR or QACT Step4, Digest the hybrid molecules by TfiI, the secondary enzyme and self-ligation Purpose, To make the hybrid molecules more small and facilitate the intramolecular ligation(TfiI end joining). The circular molecule will act as the template of inverse PCR. 4 Step5, Inverse PCR Purpose, Using the primers that locate at the HS26 fragment(Known part of the hybrid fragment) to amplify the ACPs(unknown part of the hybrid fragment) Step6, Replace the HS26 fragment at one side of the PCR product by NcoI adapor Purpose, NcoI adapor includes MmeI recognition site and MmeI can excise 19/20bp ACPs through recognize its binding site at the NcoI adapor. Step7, MmeI digestion Purpose, Remove unnecessary sequence except 19/20 bp short tags that are together with NcoI adaptor. Step8, Adding NN adaptor to the cohesive end generated by MmeI digestion Purpose, Facilitate the amplification of the tags. Step9, Amplification of the tags by PCR and excise the 19/20bp fragment by BamHI and PstI digestion. Purpose, amplifying and releasing the tags from the adapor-tag-adapor structure and facilitate the self ligation of these tags (concatemers) Step10, Self ligation of the tags to concatemers and sequencing. Purpose, it is more efficient to sequence the concatemers to get the sequence information of tags. 5