PRINCIPLE:
advertisement

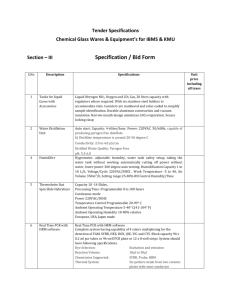
MAINE STATE POLICE CRIME LABORATORY: FORENSIC BIOLOGY SECTION AmpFlSTRSTR DNA Amplification & Typing PRINCIPLE: This procedure uses the Perkin Elmer AmpFlSTR Profiler Plus or AmpFlSTR COfiler PCR Amplification and Typing Kit. These PCR-dependent kits use Short Tandem Repeat (STR) typing technology that detects length polymorphisms. Profiler Plus detects the gender marker Amelogenin and the following nine STR loci: D3S1358, D5S818, D13S317, D7S820, D8S1179, D18S51, D21S11, FGA, and vWA. COfiler detects the gender marker Amelogenin and the following six STR loci: D3S1358, D7S820, D16S539, CSF1PO, TH01, and TPOX. When Profiler Plus and COfiler kits are combined, all thirteen CODIS core STR loci are amplified with two amplifications with two overlapping loci: D3S1358 and D7S820 (and Amelogenin). This procedure begins with PCR amplification of the loci in each multiplex. Detection of the PCR product is then performed by capillary electrophoresis of the PCR product combined with an internal size standard through the ABI Prism 310 Genetic Analyzer. The AmpFlSTR Allelic Ladders provide allele size standards for most of the alleles at each locus. SPECIMEN: The recommended range of input DNA is 0.5 to 1.5 ng. More DNA can be used if the sample is degraded. Less DNA can be used if sample size is very limited or PCR inhibitors are present. REAGENTS AND SPECIAL SUPPLIES: AmpFlSTR Profiler Plus and COfiler PCR Amplification Kits (Perkin Elmer) Hi-Di Formamide (Perkin Elmer) GeneScan-500 [ROX] Internal Lane Size Standard Kit (Perkin Elmer) 10X Genetic Analyzer Buffer with EDTA (Perkin Elmer) Milli-Q Water Performance Optimized Polymer 4 (POP-4) (Perkin Elmer) TE-4 Buffer (10 mM Tris-HCl, 0.1 mM EDTA, pH 8.0) INSTRUMENTATION AND EQUIPMENT: Collection Software v 1.0.2, or greater GeneScan Software v 2.1, or greater Heat Block Perkin Elmer 310 Genetic Analyzer 1 Version 3.2 03/13/03 MAINE STATE POLICE CRIME LABORATORY: FORENSIC BIOLOGY SECTION AmpFlSTRSTR DNA Amplification & Typing Perkin Elmer 9600 or 9700 Thermal Cycler 2 Version 3.2 03/13/03 MAINE STATE POLICE CRIME LABORATORY: FORENSIC BIOLOGY SECTION AmpFlSTRSTR DNA Amplification & Typing QUALITY ASSURANCE: 1. Ensure that all reagents satisfy the minimum standards for quality control. 2. Ensure that all equipment satisfies the minimum standards for quality control, where appropriate. 3. Wear gloves at all times during the PCR setup and typing procedures to reduce the risk of contamination. 4. Use PCR setup pipettes and aerosol resistant pipette tips to minimize contamination. 5. In order to prevent contamination, all steps in this procedure should be conducted in the proper laboratory hoods when appropriate. 6. Any laboratory workspace and all pipettes and racks to be used in this procedure must be cleaned with 10% bleach and thoroughly dried before beginning. When using a laminar flow hood, turn on ultraviolet light for a minimum of 10 minutes before and after each use. 7. To prevent contamination, microcentrifuge tubes must be irradiated in an ultraviolet crosslinker with 2 j/cm2. The extraction buffer and TE buffer (in 50 ml conical tubes) must be irradiated with 6 j/cm2. Follow "Irradiation of Reagents and Supplies in the Ultraviolet Crosslinker" SOP for appropriate method of irradiation. 8. Any equipment taken from a post-amplification room to a pre-amplification room must be sterilized with 10% bleach before removal from the post- amplification room and again in the pre-amplification room before use. Reagents and PCR product should never be transferred from a post-amplification room to a pre-amplification room. 9. Only one evidence specimen will be open at any one time. 10. Change pipette tips between each transfer or addition of sample or reagent, unless otherwise noted. 11. No aliquot of any reagent may be returned to the original stock container. (Exception: ROX/DiFormamide may be returned to original stock container from a sterile vessel.) 12. Extraction Reagent Blank Control: An extraction reagent blank control must be amplified and analyzed at least once. The volume of reagent blank amplified must be no less than the volume of sample amplified. This control will regulate for the presence of extraction contamination. The reagent blank 3 Version 3.2 03/13/03 MAINE STATE POLICE CRIME LABORATORY: FORENSIC BIOLOGY SECTION AmpFlSTRSTR DNA Amplification & Typing should be the first tube setup during amplification. The extraction reagent blank must be carried through the remaining analyses. 13. Positive PCR Control: A known positive control (Control DNA 9947A) must be included with each set of amplification reactions. The positive control should be setup as the second to last tube. All other tubes should be setup and closed prior to adding the positive control DNA. The positive control must be carried through the remaining analyses. 14. Negative PCR Control: A negative control must be included with each set of amplification reactions. The negative control should be setup last. The negative control must be carried through the remaining analyses. 15. Record all information on the STR Amplification worksheet. SAFETY: 1. Material Safety Data Sheet for all reagents used within this protocol must be read prior to amplification and typing. 2. Biological Safety Cabinets contain ultraviolet lamps. Skin and eyes should not be exposed to ultraviolet light. Wear UV protective eyeglasses when working in the vicinity of ultraviolet light. 3. The heating plate on the thermal cycler as well as the heat block can become very hot. Do not touch the heating plate or heat block while the instrument is turned on. 4. Electrical Shock Hazard: The 310 Genetic Analyzer contains a high voltage power supply. Although the instrument has been designed with safety features in the door to disconnect the power supply when the door is open, please follow procedures as prescribed. 5. Laser Hazard: The 310 Genetic Analyzer contains a laser. Operating this instrument without due regard to precautions, or in any manner not prescribed here, may be unsafe and can cause physical harm from radiation exposure. 6. Chemical Hazard: Formamide is a teratogen and is harmful by inhalation, skin contact and ingestion. Use in well ventilated area. Use chemical resistant gloves and safety glasses when handling. 4 Version 3.2 03/13/03 MAINE STATE POLICE CRIME LABORATORY: FORENSIC BIOLOGY SECTION AmpFlSTRSTR DNA Amplification & Typing PROCEDURAL NOTE: 1. If the presence of a PCR polymerase inhibitors is suspected: the sample can be re-extracted with 5% chelex, and/or centriconed, and/or a smaller aliquot of the extract can be amplified again. Additionally, 8 g/100 l of bovine serum albumin (BSA) and 1 l of AmpliTaq Gold Polymerase (5 units) can be added to the sample. If BSA and AmpliTaq Gold Polymerase are added to a sample, the proper controls (reagent blank, positive control and negative PCR control) must also be run with the added BSA and AmpliTaq Gold Polymerase. The scientist will use his/her assessment of the problem to determine the most appropriate course of action. 2. If samples have been stored on Chelex beads for a long period of time, it is advisable to boil the sample again in a water bath for 8 minutes prior to setting up PCR amplification. PROCEDURE: PCR Setup 1. Ensure that the thermal cycler is programmed with the following four linked files: 95oC for 11 min a. One-Temperature Hold File: b. Three-Temperature Cycling File: 94oC for 1 min (denature) [Total of 28 Cycles] 59oC for 1 min (anneal) 72oC for 1 min (extend) 2. c. One-Temperature Hold File: 60oC for 45 min d. One-Temperature Hold File: 4oC for forever PCR setup is performed in a laminar flow hood in a PCR product free room. Ensure that the setup area including equipment and gloves are thoroughly cleaned with 0.525% sodium hypochlorite (10% bleach). Expose the laminar flow hood to UV light for a minimum of 10 minutes. 3. Label 0.2 ml tubes with the kit name (“plus” or “cofiler”), case number, and specimen number, or derivation thereof. 5 Version 3.2 03/13/03 MAINE STATE POLICE CRIME LABORATORY: FORENSIC BIOLOGY SECTION AmpFlSTRSTR DNA Amplification & Typing 4. Vortex the tubes of AmpFlSTR PCR Reaction Mix, appropriate AmpFlSTR Primer Set, AmpliTaq Gold DNA Polymerase, and AmpFlSTR Control DNA 9947A (0.1 ng/l) for approximately 5 seconds. Spin tubes briefly in microcentrifuge. 5. Master Mix is prepared for 25 l reactions. Prepare a Master Mix by combining the following reagents in a 1.5 ml tube: a. (# of samples) x (10.5 l of AmpFlSTR PCR Reaction Mix) b. (# of samples) x (0.5 l of AmpliTaq Gold DNA Polymerase) c. (# of samples) x (5.5 l of AmpFlSTR Primer Set) 6. Mix the Master Mix thoroughly by vortexing, then spin briefly in microcentrifuge. 7. Add TE-4 to each tube so that the volume of TE-4 and DNA will be 10 l total. Pipette tip does not have to be changed between additions. 8. Add 15 l of AmpFlSTR PCR Master Mix to each tube. Pipette tip does not have to be changed between additions for a particular multiplex. 9. Add the appropriate volume of extract or control DNA to the reagent blank, specimen extract, and PCR positive control tubes. Only open one sample at a time. Cap tubes immediately after each addition. 10. Carry tubes to the PCR room and place them in the thermal cycler. If the thermal cycler is not available, the reactions can be stored at room temperature in a dark place, such as a drawer, for several hours. PCR Amplification Takes approximately 2 ½ hours. For model 9600: 1. Turn on the thermal cycler (can be done before setting up PCR, so the heated lid can reach temperature). 2. Place tubes in thermal cycler and close lid. 6 Version 3.2 03/13/03 MAINE STATE POLICE CRIME LABORATORY: FORENSIC BIOLOGY SECTION AmpFlSTRSTR DNA Amplification & Typing 3. If “RUN” is the underlined option, press [enter] (change by pressing [option]). 4. Screen will say “Run, Enter program # __”, press [3] and [enter]. 5. Make certain screen says “Select tube Micro, Reaction vol? 25ul”, and press [enter]. Screen will say “Close and tighten the sample cover” and then begin cycling. 6. Tubes may be removed when the program is done cycling and is going to, or has reached, the 4oC hold. Press [Stop] three times to end the run. 7. Place the tubes in the 4oC refrigerator for storage. Once the final case report has been administratively reviewed, the PCR product may be discarded. For model 9700: 1. Turn on the thermal cycler (can be done before setting up PCR, so the heated lid can reach temperature). 2. Place tubes in thermal cycler and close lid. 3. Press [F1] = Run. 4. Press up or down arrows to highlight “ampflstr – 28 cycle” program name. 5. Press [F1] = Start 6. Make certain screen says “Reaction volume: 25ul, Ramp speed: 9600”. 7. Press [F1] = Start. Screen will show graph of times and temperatures, flashing the part of graph that is currently being executed. 8. Tubes may be removed when the program is done cycling and is going to, or has reached, the 4 oC hold. Press [Stop] two times and [F5] = Exit to end the run. 9. Place the tubes in the 4 oC refrigerator for storage. Once the final case report has been administratively reviewed, the PCR product may be discarded. Setting Up a Run on 310 Genetic Analyzer Change gentetic analyzer buffer in autosampler vial and anode buffer reservoir every two days. Be certain the polymer in the syringe and block is not more than 5 days old. After 5 days, urea decomposition in the polymer causes transient current increases (spikes) during electrophoresis. 7 Version 3.2 03/13/03 MAINE STATE POLICE CRIME LABORATORY: FORENSIC BIOLOGY SECTION AmpFlSTRSTR DNA Amplification & Typing Allow the polymer to equilibrate to room temperature before adding polymer to the syringe. Mix polymer thoroughly by gently inverting the bottle to ensure that all precipitate goes into solution (avoid creating bubbles in the bottle of polymer) See the sections titled “Setting Up the Instrument” and “Setting Up a Run” in the SOP “Operation and Quality Assurance of the ABI Prism 310 Genetic Analyzer” to see how to properly prepare the instrument for each run. Preparing Samples and Allelic Ladders 1. Fill out form “AmpFlSTR DNA Typing Injection List.” 2. Prepare the deionized formamide and GeneScan-500 [ROX] Internal Lane Size Standard as a master mix. Combine 1.20 ml deionized formamide + 25 l GeneScan-500 [ROX] size standard. Vortex to mix, then spin briefly in a microcentrifuge. Can be stored at 2-6oC for up to two weeks. 3. Aliquot 25 l of formamide/ROX master mix into 0.2 ml strip tubes, one tube for each amplified sample, plus additional tubes for ladders. Strip tubes need not be labeled. (Alternatively, 0.5 ml Genetic Analyzer sample tubes can be used, but these tubes must be individually labeled with case number, item number, and multiplex name). 4. Add 1.5 l PCR product of amplified samples and amplification controls to the corresponding tubes containing formamide/ROX master mix. Mix by pipetting up and down or by stirring with pipette tip. 5. Add 1.5 l allelic ladder to the corresponding tubes containing formamide/ROX master mix, then mix by pipetting up and down or by stirring with pipette tip. 6. Seal the tubes with a septum strip and denature by placing at 95oC in a heat block or thermal cycler for 3 minutes. DO NOT close the thermal cycler during this step. 7. Transfer tubes to an ice water bath and chill tubes for at least 3 minutes. 8 Version 3.2 03/13/03 MAINE STATE POLICE CRIME LABORATORY: FORENSIC BIOLOGY SECTION AmpFlSTRSTR DNA Amplification & Typing Loading the Samples 1. Place the strip tubes in a 96-well retainer rack and lock the tubes in place with a retainer clip. 2. Place the rack (not the brown base) in the autosampler tray with well A1 in the right rear corner of the autosampler. 3. If a new capillary has been installed, select “Change Capillary” in the Instrument window. Select “reset counter” and “OK” in the Reset window to set the injection counter to zero. 4. Place the sample tray in the instrument. Check that the polymer lot number and expiration date is correctly entered on the 310 Log sheet on the front of the instrument and in the 310 Collection program. NOTE: If the electrode is changed or adjusted, choose “Autosampler Calibrate” from the Instrument menu before inserting the sample tray. See details in the “Operation and Quality Assurance of the ABI Prism 310 Genetic Analyzer” SOP. Preparing the Sample Sheet 1. Set up the Sample Sheet by choosing “New...” from the File menu and clicking on the “96tube GeneScan Sample Sheet” icon. A blank GeneScan sample sheet will appear 2. Fill in the sample names in the “Sample Name” column. The number of the sample (i.e. A1, A2, A3,..) corresponds to the samples’ position in the autosampler tray. 3. Copy and paste contents of “Sample Name” column into “Sample Info” column and “Comments” column, or type in identifiers such as names or descriptions of items. The word “ladder” must appear in the “Sample Info” in order for the Genotyper program to be able to locate the allelic ladder. NOTE: Descriptions typed in “Sample Info” will appear on the electropherograms. Descriptions typed in “Comments” will appear on Tables. 4. In the “Std” (Standard) column, click next to the color that represents the GeneScan standard (“R” for ROX in this case), if not already selected. 5. In the “Pres” (Present) column, be certain the check boxes for all four colors are selected. 6. Save sample sheet as the default name. 9 Version 3.2 03/13/03 MAINE STATE POLICE CRIME LABORATORY: FORENSIC BIOLOGY SECTION AmpFlSTRSTR DNA Amplification & Typing Preparing a GeneScan injection list 1. Open Collection software. Choose New from the File menu. 2. Click the “GeneScan Injection List” icon. 3. Click the arrow in the Sample Sheet field to display a pop-up menu of stored Sample Sheets. Select the Sample Sheet just created. NOTE: The default setting for the Injection List assumes that each sample will be injected once in the order listed in the Sample Sheet. If a different injection order or multiple injections are required, use the copy, insert, and paste functions to rearrange or duplicate samples in the injection list. 4. Set the desired injection time (default time is 5 seconds). Injection time can be varied from 2 seconds to 10 seconds at the analyst’s discretion. 5. Select the appropriate internal lane size standard (GeneScan-500) if not already selected. 6. Select module GS STR POP-4 (1 ml) F for every injection, if not already selected. 7. Change the run time to 26 minutes. 8. Select the appropriate Filter Set F Matrix file for every injection, if not already selected. 9. Click the Run button. Long term storage and cleaning of 310 1. If the capillary is removed from the instrument (and has been used for fewer than approximately 150 injections), fill a graduated cylinder with water, seal with a septum, and store the capillary with its ends poked through the septum and submerged in the water. Do not allow the ends of the capillary (and the polymer inside) to dry out. 2. Discard unused polymer if it has been in the instrument more than 5 days. Remove and clean the syringe, anode buffer reservoir, and block with warm water, rinse with Milli-RO water, and air dry. 3. Remove buffer, water, and waste vials from autosampler, rinse with water, and air dry. Automated Analysis Follow SOP “STR Analysis and Interpretation” for analysis and interpretation of STR profiles. 10 Version 3.2 03/13/03 MAINE STATE POLICE CRIME LABORATORY: FORENSIC BIOLOGY SECTION AmpFlSTRSTR DNA Amplification & Typing REFERENCES: ABI Prism™ 310 Genetic Analyzer User’s Manual, Rev. 1, Perkin Elmer Corp., July 1995. ABI Prism™ GeneScan® Analysis 2.1 User’s Manual, Perkin Elmer Corp., September 1996. AmpFlSTR Profiler™ PCR Amplification User’s Manual, Ver. A, Perkin Elmer Corp., 1997. AmpFlSTR Profiler Plus™ PCR Amplification User’s Manual, Ver. A, Perkin Elmer Corp., 1997. Reviewed Date Supervisor, Forensic DNA Section Adopted Date Director, Crime Laboratory Annual Review: Reviewed Date Reviewed Date Reviewed Date Reviewed Date Reviewed Date Reviewed Date Reviewed Date Reviewed Date Reviewed Date 11 Version 3.2 03/13/03 MAINE STATE POLICE CRIME LABORATORY: FORENSIC BIOLOGY SECTION AmpFlSTRSTR DNA Amplification & Typing Reviewed Date 12 Version 3.2 03/13/03