ch20
advertisement

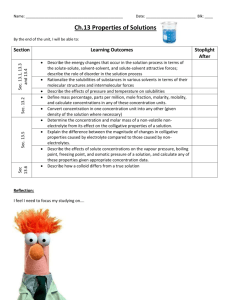
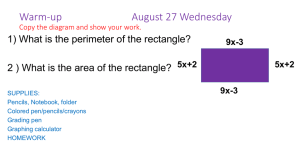
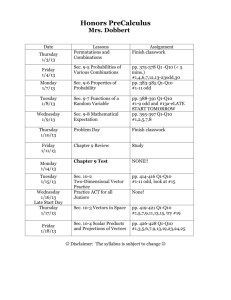
Chapter 20 Nucleic Acids Multiple Choice 1. Which structures are named correctly? (Sec. 20.2) O O O a) b) c) d) CH3 HN HN O N O NH2 N N N N N N HN NH2 N H H H H guanine I thymine II N adenine III uracil IV I, II II, III III, IV I, III 2. Which structure is a nucleoside? (Sec. 20.2) O NH2 N N b) a) N HOH2C O N O N H O O O CH3 HN HOH2C O HOH2C N d) O O O OH OH O c) CH3 HN OH P O O 247 OH P O O Chapter 20 Nucleic Acids 3. Which structures are named correctly? (Sec. 20.2) O NH2 CH3 HN HOH2C O N HOH2C O O O O CH3 N N O OH O P O O O OH P O O O P O O thymidine 5'-monophosphate I cytidine 3'-diphosphate II O NH2 N HN O O P OH2C H2N O O OH N N H O O O P O P OH2C O O OH a) b) c) d) N O OH adenosine 5'-monophosphate III N N N H OH adenosine 5'-diphosphate IV I, II II, III I, IV II, IV 248 Chapter 20 Nucleic Acids 4. Which nucleotides are named correctly? (Sec. 20.2) O HN O O N O O OH P OH2C H2N O O OH OH O 5'-TMP I CH3 HN HOH2C O O O O OH N H OH 3'-GMP II NH2 O O P O P OH2C O O N N N O OH OH P O 5'-GDP IV O 3'-TDP III I, III II, III III, IV II, IV 249 N H P O O O N N O a) b) c) d) N HN O P OH2C O O O CH3 Chapter 20 Nucleic Acids 5. What is the correct name for this structure? (Sec. 20.2) O CH3 HN HOH2C O N O O O P O O a) b) c) d) 5’-dAMP 5’-TMP 3’-AMP 3’-dTMP 6. How do DNA and RNA differ? (Sec. 20.3, 20.4) I) the position of attachment of phosphate groups II) the position of attachment of base groups III) the sugar structure at C-2’ IV) structure of bases a) b) c) d) I, II II, III III, IV II, IV 7. Which are the correct pairings of bases in the structure of DNA? (Sec. 20.3) I) A-C II) G-T III) A-G IV) G-C V) A-T VI) C-T a) b) c) d) I, II III, V III, IV IV, V 250 Chapter 20 Nucleic Acids 8. In both RNA and DNA structures, the phosphate groups are attached to the sugar structure at which positions? (Sec. 20.3, 20.4) a) b) c) d) 5’, 5’ 3’, 3’ 3’, 5’ 5’, 3’ 9. What is the charge on adenosine phosphate at blood serum pH of 7.4? (Sec. 20.3) a) b) c) d) 0 –1 –2 –3 10. Which is the complementary tetranucleotide for 5’-AGCT-3’? (Sec. 20.3) a) b) c) d) 5’-TCGA-3’ 5’-CTAG-3’ 5’-AGCT-3’ 5’-GATC-3’ 11. How many nucleotides direct a specific amino acid into a polypeptide sequence? (Sec. 20.5) a) b) c) d) 1 2 3 4 12. Which is the DNA complement for 5’-ACCGTTAAT-3’? (Sec. 20.3) a) b) c) d) 5’-ATTAACGGT-3’ 5’-GTTACCGGC-3’ 5’-TGGCAATTA-3’ 5’-CAATGGCCG-3’ 251 Chapter 20 Nucleic Acids 13. Hydrogen bonding is strongest between which two bases? (Sec. 20.3) NH2 O N O a) b) c) d) CH3 HN N O NH2 N N N H H I II O N N N HN NH2 N H III N H IV I, II I, IV II, III II, IV 14. Where does protein synthesis occur? (Sec. 20.4) a) b) c) d) chromatin ribosome codon histones 15. Which is the complementary sequence in RNA for the DNA sequence 5’-AATCAGTT-3’? (Sec. 20.4) a) b) c) d) 5’-AAUCAGUU-3’ 5’-AACUGAUU-3’ 5’-UUAGUCAA-3’ 5’-CCAUCGAA-3’ 16. A complete turn of the DNA helix occurs at which distance? (Sec. 20.3) a) b) c) d) 34 Å 10 Å 20 Å 4Å 17. Which statement about the base mole-percent composition of DNA is true? (Sec. 20.3) a) b) c) d) A=G=C=T (A+G)=(C+T) (A+T)=(G+C) There is no relationship 18. The backbone of DNA is best described as: (Sec. 20.3) a) b) c) d) pairs of complementary bases poly(1,3-diol-phosphate esters) pyrophosphates sugar glycosides 252 Chapter 20 Nucleic Acids 19. Which statements about differences between DNA and RNA are true? (Sec. 20.3, 20.4) I) The sugar unit is -D-ribose in RNA and -2-deoxyribose in DNA. II) The pyrimidine bases are uracil and thymine in RNA and cytosine and thymine in DNA. III) RNA is single stranded, DNA double stranded. IV) The phosphodiester groups join the 3’ to 5’ ends of sugars in RNA, and join the 5’ to 3’ ends in DNA. a) b) c) d) I, II I, III II, III III, IV 20. Which are types of RNA? (Sec. 20.4) I) ribosomal II) histonal III) helix IV) transfer a) b) c) d) I, II I, III II, IV I, IV 21. Which is the order of increasing percentage in cells of messenger RNA, transfer RNA, ribosomal RNA (least first)? (Sec. 20.4) a) b) c) d) mRNA, tRNA, rRNA tRNA, rRNA, mRNA rRNA, tRNA, mRNA mRNA, rRNA, tRNA 22. Which statements about mRNA codons are correct? (Sec. 20.4) I) Each amino acid is coded by only one codon. II) Codons code for several amino acids. III) UAA, UAG, UGA are stop codons. IV) Several amino acids are coded by more than one codon. a) b) c) d) I, II III, IV I, III II, IV 23. Which is the key to the chain termination method of DNA sequencing? (Sec. 20.6) a) b) c) d) polyacrylamide gels restriction endonucleases primer 2’,3’-dideoxynucleoside triphosphate 253 Chapter 20 Nucleic Acids 24. Which is the key to in vitro DNA replication? (Sec. 20.6) a) b) c) d) polyacrylamide gels restriction endonucleases primer 2’,3’-dideoxynucleoside triphosphate 25. Which statements about restriction endonucleases are true? (Sec. 20.6) I) Each cleave DNA at the same site, but leave different fragment lengths. II) Each recognizes 4 to 8 nucleotides, and cleaves at all sites containing the sequence. III) Approximately 1000 restriction endonucleases have been isolated and characterized. IV) Each cleaves the DNA strand by reducing the base off at the sugar 1’ position. a) b) c) d) I, IV II, IV II, III III, IV Fill in the Blank 1. The name of the following base is ____________________________. (Sec. 20.2) O HN O N H 2. The name of the following nucleoside is _____________________________________. (Sec. 20.2) O N HN H2N N N CH2OH O OH OH 254 Chapter 20 Nucleic Acids 3. The name of the following nucleotide is _____________________________________. (Sec. 20.2) NH2 N O O O P O CH2 O N O OH OH 4. The complementary DNA single strand of 5’-AGTGTTCCAGT-3’ is 5’-____________________________-3’. (Sec. 20.3) 5. The complementary RNA strand to the DNA single strand 5’-GGACTTGACTA-3’ is 5’-____________________________-3’. (Sec. 20.4) 6. The first four amino acids of the protein coded for by AUUAUCCAUAAUGGUGGAAAACGA are __________________________________________________________. (Sec. 20.5) 7. The first four amino acids of the protein coded for by GCUGCCGAUGACGGACGGUGGUGU are __________________________________________________________. (Sec. 20.5) 8. An mRNA strand that codes for the pentapeptide Ile-Thr-Pro-Lys-Arg could have __________ codons in the first position. (Sec. 20.5) 9. An mRNA strand that codes for the pentapeptide Met-Ser-Tyr-Met-Cys could have __________ codons in the first position. (Sec. 20.5) 10. The original DNA strand that results in the following fragments after the Dideoxy method is 5’-_____________________________-3’. (Sec. 20.6) Primer = AGCT Fragments AAGCT, GAAGCT, AGAAGCT, AAGAAGCT, CAAGAAGCT, TCAAGAAGCT True-False 1. The following nucleoside is named Cytidine. (Sec. 20.2) NH2 N O O O P O CH2 O OH N O OH 255 Chapter 20 Nucleic Acids 2. The following nucleotide is named 5’-Guanisine diphosphate. (Sec. 20.2) O N HN P O H2N H2 P O C O O O O O N N O OH OH 3. The mole % of purine bases must be equal in human DNA. (Sec. 20.3) 4. The tertiary structure of DNA involves supercoiling. (Sec. 20.3) 5. t-RNA is the lowest molecular weight and most abundant of the RNAs. (Sec. 20.4) 6. mRNA is the shortest lived of the RNAs and the least abundant. (Sec. 20.4) 7. The complementary strand of mRNA to the DNA sequence TAGAAGTAG is ATCTTCATC. (Sec. 20.4) 8. tRNA with the codon UGA codes for Cys. (Sec. 20.5) 9. If the smallest fragment produced during the Dideoxy method and separated by PAGE includes the primer and the base T, the original DNA template begins with T after the primer sequence. (Sec. 20.6) 10. The Chain Termination method uses dideoxy nucleosides to stop DNA synthesis. (Sec. 20.6) 256 Chapter 20 Nucleic Acids Answers Multiple Choice 1. b 2. c 3. d 4. a 5. d 6. c 7. d 8. c 9. c 10. c 11. c 12. a 13. b 14. b 15. b 16. a 17. b 18. b 19. b 20. d 21. a 22. b 23. d 24. c 25. c Fill in the Blank 1. Thymine 2. Guanisine 3. 5’-cytidine monophosphate 4. 5’-ACTGGAACACT-3’ 5. 5’-UAGUCAAGUCC-3’ 6. Ile-Ile-His-Asn 7. Ala-Ala-Asp-Asp 8. 3 9. 1 10. 5’-AGTTCT-3’ True-False 1. F 2. T 3. F 4. T 5. F 6. T 7. F 8. F 9. F 10. T 257