R script to impute missing loci by population.

R scrpt to impute missing SNP loci by population- Matthew H. Grinnell
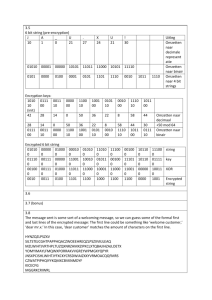
The script ImputMissing.R imputes missing genetic data using the mean of the concerned allele, by population. Currently, this script works for SNP data, but not for microsatellite data. This script uses the R package adegenet (Jombart and Ahmed, 2011). First, copy the genepop _le to the Data folder in the work- ing directory (i.e., C:/Grinnell/Workspace/Genetics/Data). Then, source the script and choose the genepop _le name interactively, such as Eulachon.gen, when prompted. The default location to select the genepop _le, and save the imputed genepop _le (e.g., Eulachon.Fill.gen), is the aforementioned direc- tory. The following genepop _le with three populations and 7 SNPs has missing loci in multiple locations.
###############################################################################
#
# Author: Matthew H. Grinnell
# Affiliation: Pacific Biological Station, Fisheries and Oceans Canada (DFO)
# Group: Molecular Genetics Lab
# Address: 3190 Hammond Bay Road, Nanaimo, BC, Canada, V9T 6N7
# Contact: e-mail: matt.grinnell@dfo-mpo.gc.ca | tel: 250.756.3367
# Project: Genetics
# Code name: ImputeMissing.R
# Version: 1.0
# Date started: Sep 11, 2014
# Date edited: Jan 05, 2015
#
# Overview:
# Impute missing values in genepop files, using the loci mean, by population.
#
# Requirements:
# A genepop file (i.e., /Data/*.gen).
#
# Notes:
# Creates a new genepop file (i.e., /Data/*.Fill.gen). Simply use the function
# 'na.replace' in the package 'adegenet' if you want to impute missing values
# using the loci mean over all populations (i.e., instead of imputing using the
# loci mean by population).
#
###############################################################################
########################
##### Housekeeping #####
########################
# General options rm( list=ls( ) ) # Clear the workspace sTime <- Sys.time( ) # Start the timer
graphics.off( ) # Turn graphics off
# Install missing packages and load required packages (if required)
UsePackages <- function( pkgs, update=FALSE, locn="http://cran.rstudio.com/" ) {
# Identify missing (i.e., not yet installed) packages
newPkgs <- pkgs[!(pkgs %in% installed.packages( )[, "Package"])]
# Install missing packages if required
if( length(newPkgs) ) install.packages( newPkgs, repos=locn )
# Loop over all packages
for( i in 1:length(pkgs) ) {
# Load required packages using 'library'
eval( parse(text=paste("library(", pkgs[i], ")", sep="")) )
} # End i loop over package names
# Update packages if requested
if( update ) update.packages( ask=FALSE )
} # End UsePackages function
# Make packages available
UsePackages( pkgs=c("adegenet") )
####################
##### Controls #####
####################
# Name of the input mixture file (*.mix) S:/*.* inputFN <- choose.files( default=file.path(getwd(), "Data/*.*"),
caption="Pleas select a genepop file", multi=FALSE )
# Name of the output mixture file outputFN <- paste( strsplit( x=inputFN, split=".gen" )[[1]], ".Fill.gen",
sep="" )
# Population identifier popID <- "POP"
################
##### Data #####
################
# Read genetic data dat <- read.genepop( file=inputFN, quiet=TRUE, missing=NA )
# Scan the first line firstLine <- readLines( con=inputFN, n=1 )
################
##### Main #####
################
# Stop if any sample names are repeated (i.e., non-unique) if( any(duplicated(dat@ind.names)) )
stop( "Non-unique sample name(s): sample(s) ",
which(duplicated(dat@ind.names)), call.=FALSE )
# Fill in missing data using mean by population
FillMissingByPop <- function( gen, imputeMeth="mean" ) {
# Convert all populations to a data frame
genDF <- genind2df( x=gen )
# Split the data frame by population
dfSplit <- split( x=genDF, f=genDF$pop )
# Message
cat( "Input file '", inputFN, "' has ", length(gen@loc.names), " loci, ",
length(gen@pop.names), " populations, and ", length(gen@ind.names),
" samples:\n", sep="" )
# Write the first line
write( x=paste(firstLine, "(with imputed data)"), file=outputFN )
# Write the loci names
write( x=gen@loc.names, file=outputFN, append=TRUE )
# Loop over populations
for( i in 1:length(dfSplit) ) {
# Get the ith population
iDF <- dfSplit[[i]]
# Convert to genind (only one population)
iGenind <- df2genind( X=subset(iDF, select=-pop), type="codom" )
# Add population information
iGenind@pop.names <- as.character( unique(iDF$pop) )
# Print message
cat( "\tPopulation ", iGenind@pop.names, " with ", nrow(iDF),
" samples: imputed ", sum(is.na(iDF)), " missing values via '",
imputeMeth, "'\n", sep="" )
# Fill missing data using the mean
iGenFill <- na.replace( x=iGenind, method=imputeMeth, quiet=TRUE )
# Convert to data frame
iDFFill <- genind2df( x=iGenFill, pop=iDF$pop )
# Append population to a new data frame
ifelse( i==1,
dfFill <- iDFFill,
dfFill <- rbind(dfFill, iDFFill) )
# Write the population marker
write( x=popID, file=outputFN, append=TRUE )
# Get the data for the output
outDat <- subset( iDFFill, select=-pop )
# Modifty row names for output
rownames( outDat ) <- paste( rownames(outDat), "," , sep="" )
# Write the genetic info
write.table( x=outDat, file=outputFN, append=TRUE, quote=FALSE,
col.names=FALSE )
} # End loop over populations
# Convert all populations to genind object
genFill <- df2genind( X=subset(dfFill, select=-pop), pop=dfFill$pop )
# Return the filled data set
return( genFill )
} # End FillMissingByPop function
# Fill missing data datFill <- FillMissingByPop( gen=dat )
###############
##### End #####
###############
# Print end of file message and elapsed time cat( "End of file ImputeMissing.R: ", sep="" ) ; print( Sys.time( ) - sTime )
Example data set
Eulachon significant SNPs
Tpa_0012
Tpa_0024
Tpa_0035
Tpa_0226
Tpa_0237
Tpa_0269
Tpa_0342
POP
TpacTMR01-003, 0000 0102 0101 0101 0202 0102 0102
TpacTMR01-004, 0102 0101 0101 0101 0000 0101 0102
TpacTMR01-005, 0102 0101 0101 0102 0202 0101 0102
POP
TpacKEN04-003, 0101 0101 0101 0101 0102 0101 0102
TpacKEN04-006, 0000 0101 0101 0101 0000 0102 0202
TpacKEN04-007, 0101 0101 0101 0102 0000 0101 0000
TpacKEN04-008, 0101 0101 0101 0000 0101 0101 0102
POP
TpacSTIK06-002, 0000 0101 0101 0000 0102 0101 0102
TpacSTIK06-004, 0102 0101 0101 0000 0000 0102 0101
TpacSTIK06-010, 0102 0101 0101 0101 0000 0101 0101
TpacSTIK06-012, 0000 0102 0101 0102 0102 0101 0101
TpacSTIK06-013, 0101 0000 0000 0101 0000 0102 0101