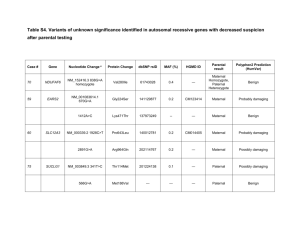
Table S1. Summary statistics for microsatellites used for pedigree
advertisement

Table S1. Summary statistics for microsatellites used for pedigree creation and adoption detection. E1 = allelic dropout rate, E2 = false allele rate, NA = number of alleles, HO = observed heterozygosity, HE = expected heterozygosity, Pex = probability of exclusion for a single unknown parent, PID = probability of identity for unrelated individuals. For details on how each of these numbers was calculated, consult the main manuscript. Locus CXX20 CXX110 CXX173 G1A G10B G1D G10L G10C G10M G10P G10X UarMU05 UarMU10 UarMU23 UarMU26 UarMU50 UarMU51 UarMU59 G10H G10J G10U MSUT-1 MSUT-2 MSUT-6 MSUT-8 Reference Ostrander et al. (1993) Ostrander et al. (1993) Ostrander et al. (1993) Paetkau and Strobeck (1994) Paetkau and Strobeck (1994) Paetkau and Strobeck (1994) Paetkau and Strobeck (1994) Paetkau et al. (1995) Paetkau et al. (1995) Paetkau et al. (1995) Paetkau et al. (1995) Taberlet et al. (1997) Taberlet et al. (1997) Taberlet et al. (1997) Taberlet et al. (1997) Taberlet et al. (1997) Taberlet et al. (1997) Taberlet et al. (1997) Paetkau et al. (1998) Paetkau et al. (1998) Paetkau et al. (1998) Kitahara et al. (2000) Kitahara et al. (2000) Kitahara et al. (2000) Kitahara et al. (2000) E1 0 0 0.009 0 0 0 0 0 0 0 0 0 0 0 0 0 0.010 0 0 0 0.039 0 0 0 0 E2 0.012 0 0.007 0 0 0 0 0 0.013 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 NA 9 10 6 8 7 6 6 8 9 9 9 10 7 3 9 9 8 10 10 4 7 8 9 3 6 HO 0.787 0.590 0.694 0.531 0.601 0.626 0.432 0.674 0.789 0.743 0.746 0.785 0.638 0.112 0.837 0.788 0.798 0.847 0.766 0.639 0.569 0.749 0.765 0.520 0.767 HE 0.795 0.600 0.691 0.540 0.607 0.634 0.427 0.681 0.797 0.749 0.728 0.771 0.643 0.112 0.840 0.781 0.801 0.831 0.767 0.628 0.574 0.738 0.775 0.524 0.771 Pex 0.427 0.223 0.276 0.158 0.217 0.222 0.096 0.270 0.433 0.363 0.326 0.403 0.236 0.006 0.510 0.393 0.435 0.496 0.383 0.201 0.186 0.326 0.395 0.137 0.377 PID 0.070 0.180 0.141 0.252 0.184 0.182 0.356 0.146 0.068 0.093 0.114 0.077 0.176 0.791 0.045 0.082 0.067 0.050 0.086 0.209 0.215 0.113 0.082 0.298 0.088 Table S2. Summary statistics for the final pedigree, including genetic parentage assignments and maternal relationships derived from field data. All statistics were calculated using the R package PEDANTICS 1.5 (Morrissey & Wilson 2010). Records Maternities Paternities Full sibships Maternal sibships Maternal half-sibships Paternal sibships Paternal half-sibships Maternal grandmothers Maternal grandfathers Paternal grandmothers Paternal grandfathers Maximum pedigree depth Founders Mean maternal sibship size Mean paternal sibship size Non-zero F F > 0.125 Mean pairwise relatedness Pairwise relatedness ≥ 0.125 Pairwise relatedness ≥ 0.25 Pairwise relatedness ≥ 0.5 4449 2957 1861 551 6403 5852 3009 2458 1401 739 487 329 5 1381 3.66 2.81 3 0 7.90×10–4 2.94×10–3 1.72×10–3 5.43×10–4 References Kitahara, E., Y. Isagi, Y. Ishibashi and T. Saitoh. 2000. Polymorphic microsatellite DNA markers in the Asiatic black bear Ursus thibetanus. Molecular Ecology 9:1661–1662. Morrissey, M. B. and A. J. Wilson. 2010. PEDANTICS: an R package for pedigree-based genetic simulation and pedigree manipulation, characterization and viewing. Molecular Ecology Resources 10:711–719. Ostrander, E. A., G. F. Sprague and J. Rine. 1993. Identification and characterization of dinucleotide repeat (CA)n markers for genetic mapping in dog. Genomics 16:207–213. Paetkau, D., W. Calvert, I. Stirling and C. Strobeck. 1995. Microsatellite analysis of population structure in Canadian polar bears. Molecular Ecology 4:347–354. Paetkau, D., G. F. Shields and C. Strobeck. 1998. Gene flow between insular, coastal and interior populations of brown bears in Alaska. Molecular Ecology 7:1283–1292. Paetkau, D. and C. Strobeck. 1994. Microsatellite analysis of genetic variation in black bear populations. Molecular Ecology 3:489–495. Taberlet, P., J. J. Camarra, S. Griffin, et al. 1997. Noninvasive genetic tracking of the endangered Pyrenean brown bear population. Molecular Ecology 6:869–876.