File - Communication In Science
advertisement

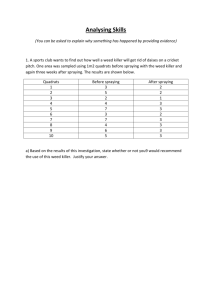
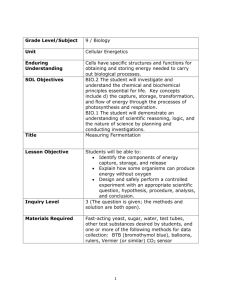
Hee Yen Zhen 42724614 MICR3003: Molecular Microbiology Practical: Beer & Biofuels Abstract Fermentation is a process where Saccharomyces cerevisiae (yeast) produces ethanol and biomass. The selected yeasts with specific strains from Wyeast Laboratories and White Lab collection were chosen in this experiment. The aim of this paper is to culture the four selected yeast strains (S288C, WLP004, WLP099 and WLP500) and undergone fermentation in yeast nitrogen base 2% glucose broth. The ethanol (biofuel) production was measured using Gas Chromatography and the biomasses are recorded using a measuring weight. The specific traits of the yeasts were determined using bioinformatics approach based on their presence of ethanol concentration and biomass production. The WLP500 yeast strains were able to produce the highest amount of ethanol concentration 1.18% (with an average of 1.12%) and WLP400 yeast strains were able to produce the most biomass, 147 mg/ml (with an average of 137.33 mg/ml). WLP099 were assumed to produce the highest ethanol and biomass production in this paper but were unsuccessful due to practical handling and human error during the experiment. In this paper, WLP500 yeast strains were selected to be the best yeast strains in the production in biofuel and Vegemite for WLP500 were able to produce high ethanol concentration and about similar biomass as WLP400 strains. Introduction Saccharomyces cerevisiae or also known as Brewer’s yeast / Baker’s yeast are single-cell eukaryotes that play an important role in our daily agriculture and food processing. Yeast can be traced back 3,000 years where the Egyptians and Babylonians utilised the fermentation of wild yeast to alveolate the bread and is then later discovered by Louis Pasteur in 1845, claimed that these microorganisms are capable of fermenting sugar, produce ethanol and carbon dioxide. Since then, Saccharomyces cerevisiae are industrialized in manufacturing fermented product. The byproduct of Saccharomyces cerevisiae from fermenting the sugar were commonly used in alcoholic beverages and fermented products in the industries, and applicable research in laboratory in searching and providing the most efficient and specific yeast for the market needs (1, 2, 3). Biofuel is a type of hydrocarbon solution that consists of ethanol for combustion in the engine of all the transportation. The amount of ethanol in the biofuel is essential and the absence of ethanol causes no combustion. Therefore, using yeast to generate biofuel from sugar is an alternative possibility. It was recently reported that University of Texas researchers were trying to convert yeast cells into generating biofuels in replacing fossil fuels(4). The selected yeasts from the list from Wyeast Laboratories and White Lab collection strains were WLP500, WLP099, WLP004 and S288C, based on each of their specific genes (ADH1, PDC1 and CDC19) were selected from the superpathway of glucose fermentation to determine which genes are responsible for the efficiency of ethanol production. The ADH1 gene is responsible in converting acetaldehyde to ethanol in the last step of fermentation pathway. The PDC1 gene is one of the pyruvate decarboxylase isoenzymes that convert pyruvate into acetaldehyde and carbon dioxide during the fermentation before the ADH1 gene. The CDC19 gene is a pyruvate kinase gene that will converts phosphoenopyruvate (PEP) for both aerobic and anaerobic respiration. Without CDC19 gene, the yeasts were not able to uptake glucose as the carbon source (5, 6, 7, 8). The microbial biomass produced from Saccharomyces cerevisiae is a suitable supplement of protein and vitamin B which were widely used in the commercial health products. The specific gene (TPK1) that attach a subunit of cAMP-dependant protein kinase A that will regulates cell growth, stress response and other processes (9, 10). In this paper, the four selected yeasts’ strains were determined the best for production in biofuel and Vegemite. The yeast strains that were able to produce high ethanol will produce less biomass, therefore WLP099 and WLP500 were proposed to produce high ethanol concentration than the other selected strains. Materials and Methods Firstly, the selected yeast namely WLP500, WLP099, WLP004 and S288C were culture into 3 falcon tubes containing 8ml yeast nitrogen base (YNB) 2% glucose each and plated on each YPD culture plates respectively and the plates were incubated at 18°C. That would be the starter cultures during that time. Once the 12 starter cultures had showed growth, each of the 12 tubes was resuspended and pitched into a 250ml bottle containing 150ml YNB individually. The bottles were labelled according to the 12 previous falcon tubes. These bottles were then capped with airlocks and incubated at 21°C for a week. As for the DNA purification, the grown yeast on the culture plates were then scraped up a large match head worth with a pipette tips and resuspended in microfuge tube contained 0.5ml TENTs buffer. The yeasts were then added with 0.2ml of acid wash 0.45mm glass beads and 0.5ml phenol-chloroform and vortex for 20 minutes. These yeasts were then centrifuged for 10 minutes at max speed which will result a multiple phase where the top clear aquas phase is extracted and transferred into a new labelled microfuge tube. 40ul (1/10v) of 3M sodium acetate and 1 ml 100% ethanol is added, then the tubes were inverted 3 times to mix. The DNA will precipitate at -20°C for about 30 minutes or more. The tubes were then centrifuged for 15 minutes at max speed. The supernatant of each tube were decanted and carefully without disrupting the pellets. The pellets were then washed with 1ml of 70% of ethanol and centrifuged for 1 minute. The supernatant is decanted and the pellets were left to dry. When the pellets were dried, 150ul TE buffer is added to dissolve the DNA and stored at -20°C for a week. Next, when all the bottles had undergone fermentation, they will be tested for Gas Chromatography (GC) for Ethanol content. The solution in the bottles was shook and about 5ml of the solution were drawn with a syringe. The syringe was then attached with a filter unit to filter out the solution into a falcon tube and 1.5ml was then pipetted into a 2ml GC sample vial. These processes were repeated for the rest of the bottles. As for the biomass measurement, 10ml of the solution was pipetted from the same bottles into a falcon tube respectively. The tubes were centrifuged and the supernatant were decanted. The samples mass were then weighted and calculated. As for the Random Amplification of Polymorphic DNA (RAPD), the isolated DNA from the microfuge tubes were resuspended and 1ul of each DNA of the yeast strain is pipetted into PCR tubes containing 24ul of PCR Master Mix and labelled respectively. The primers for this RAPD are M13 and (GACA)4 and were done separately which consist of 10 PCR tubes. The PCR will undergone 35 cycles and were stored at 4°C for a week. Finally, the PCR tubes containing the DNA of the yeasts strains were added with 2ul of loading dye individually and vortex to mix. There were 2 gels prepared for the different primers respectively. The first gel is labelled for M13 primer detection. 6ul of 1kb ladder was loaded to both ends of the wells and the stained yeast strains were pipetted accordingly (control, S288C, WLP004, WLP099 and WLP500). The process were repeated for the second gel which is labelled for (GACA)4 detection. The voltage was set to 90V and both gels were run for 30 minutes. Once the front dye had migrated about ¾ distances away from the wells, the run was stopped and the gels were placed on a UV transmilluminator to visualise the bands. The results of the gels were photographed. As for the bioinformatics approach which was done by a lab partner (Kurt Harris), the Illumina Data were imported into CLC and the S288C reference genome was downloaded using CLC. Each genome strains of the yeasts (WLP004, WLP099 and WLP500) were used to align with the S288C strains. Probabilistic Variation Detection such as finding the variants, SNPs, MNPs, and InDels were conducted and the total numbers of variants per strain were recorded. Marker genes were found using genome database and the first three key genes were selected which involved in the fermentation pathway. The numbers of variants per gene of each strain were recorded. Results Fermentation. All the starter cultures except one of the three WLP099 starter culture did not grow and the bottle that was prepared for its fermentation was used as a Control. Based on Table 1, the three bottles of yeast strains S288C did not grew in the media similarly to the Control. The other bottles for WLP004, WLP099 and WLP500 showed growth and fermentation in the bottles. There are no bubbles or carbonation occurred in the fermentation bottles for the media used in the fermentation is YNB 2% glucose. Biomass Measurement. The absence of fermentation from Control and S288C bottles would not be measured for their biomass. From Table 2, the biomass for yeast strains WLP004, WLP500 and WLP099 has been collected and calculated from 10ml samples. The average biomasses are 13.73 mg/ml, 12.73 mg/ml and 10.8 mg/ml respectively. WLP004 produced the most biomass, while WLP009 produced the least biomass showed in Table 2. Gas Chromatography. The Gas Chromatography results obtained were all calculated in Area (uV.s) within 1.997 to 2.001 minutes timeframe. The Graph 1 has been tabulated based on the standard ethanol of 2%, 4%, 6%, 8% and 10% in to measure the percentage of ethanol from 1ml of GC samples and Table 3 has been tabulated. The absence of ethanol in Control and S288C showed mild production of ethanol. The WLP500 yeast strains produced the highest ethanol concentration which is about 1.18% individually and an average of 1.12%, while the WLP099 yeast strains produced the least ethanol concentration which is about 1.06% individually and an average of 10.8%. Random Amplification of Polymorphic DNA (RAPD). The bands in Figure 1 showed that the random amplification produced by the primer (GACA) 4. The S288C had dark bands between 0.5-1.0kb and a light band below 0.5kb. The WLP004 had a dark band between 0.51.0kb. The WLP099 had a dark band below 0.5kb. The WLP500 had a dark band similar to the bands in WLP004 and S288C, and a light band below 0.5kb. The bands in Figure 2 showed that the random amplification produced by the primer M13. All the yeast strains in Figure 2 showed there were 2 dark bands between 0.5-1.0kb. The 288C and WLP099 had an additional dark band at 3.0kb, the WLP099 and WLP500 had an additional dark band between 2.0-3.0kb, and the WLP004 has a dark band at 2.0kb. The common bands that showed in all the yeast strains were tend to be the common consensus region from the yeasts’ strains Bioinformatics. From the bioinformatics approach, the Saccharomyces cerevisiae [Gene annotations (638)] was used as the reference genome to compare the yeast strains (Figure 5, 6, 7, 8). The alignment of the yeast strains (S288C, WLP004, WLP009 and WLP500) showed in Figure 3. The number of variants had been detected from the yeast strains and tabulated in Table 4. The three key genes (ADH1, PDC1 and CDC19) was selected from the Saccharomyces cerevisiae fermentation pathway (4) and another gene (TPK1) to determine the production of biomass has been tabulated in Table 5. The yeast strain, WLP500 had the highest number of variants compare to the other yeasts’ strains (Table 4). This showed that WLP500 had variants present in the 4 selected marker genes (Table 5). The yeast strains were then tabulated in a phylogenetic tree (Figure 3) to determine the likelihood among the yeast strains. The WLP004 yeast strains from Figure 3 was shown as outgroup and the furthest distance, 0.026775, between WLP004 clade and S288C clade. Discussion The initial of the experiment showed the absence of culture growth in one of the WLP099 falcon tube followed by the absence of fermentation in all S288C bottles. The experiment was then preceded to GC analysis and biomass measurement with the fermented samples. From Table 3, the individual bottle of WLP500 produced the highest concentration of ethanol, 1.18% and average of 1.12% within a week. WLP099 was assumed to produce higher amount of ethanol than WLP500 based on the information listed from Wyeast Laboratories and White Labs collection strains (Table 6), but from the results in Table 3, WLP099 yeast strains weren’t able to produce high ethanol concentration and much lower than WLP004, which were assumed to be the lowest production of ethanol in this experiment. The WLP099 biomass were assumed to produce within the range between WLP500 and WLP004 biomass production based on their flocculation provided in Table 6. However, the data collected showed in Table 2 showed WLP099 biomass was lower than WLP500 by 19.33mg/ml. These claimed that WLP099 might have undergone practical handling error which explains the absence of growth in WLP099 starter culture. Furthermore, the WLP099 yeast were unable to execute its full potential during the fermentation might due to the short timeframe given. From the bioinformatics approach, the yeast strain S288C was used as the reference yeast strain of the experiment. In Figure 1, the primer (GACA) 4 was able to fingerprint the common region of WLP500, WLP004 and S288C yeasts’ strains. This might showed that the three yeast strains had common properties such as alcohol tolerance or the limitation of attenuation (Table 6), while the WLP099 band which were lower than 0.5kb, might showed the properties of high alcohol tolerance or higher attenuation than the other yeast strains. As Figure 2, the bands produce from the primer M13 showed almost all the bands are common in all the strain except the density of the bands. The high density bands between 0.5-1.0kb showed the relativeness of S288C and WLP500 based on ADH1 alignment in Figure 3, but the band at 3.0kb showed low density which explained the variable of WLP500 in Figure 3, differed from other strains from position 623-682 bases (purple boxes). On the other hand, the bands between 2.0-3.0kb are present in S288C, WLP099 (high density) and WLP500 (low density). These might be the band of interest in producing high amount of ethanol, which the primer M13 replicated the most in WLP099 based on the high density of the band. The ADH1 alignment (Figure 3) showed at position 925 bases (red box) was Guanine for WLP099 where others were Adenine. This might conclude that the position 925 bases (red box) in WLP099 were the specific region for producing high amount of ethanol. As for CLC Genomic Workbench Approach, WLP500 had the highest number of variants detected (Table 4) which leads to the present in all selected marker genes (Table 5). This explained that the results of WLP500 had high ethanol production (Table 3) in this experiment. The S288C and WLP099 showed lower in both number of variants and marker genes detected (Table 4 & 5). These claims explained that results of absence growth during the fermentation process of this experiment. In conclusion, WLP500 was practically the best selected yeast strain for the production of ethanol and biomass which was the biofuel and Vegemite for this paper from both analysed results and the bioinformatics analysis. The WLP099 was theoretically assumed to be also the best selected yeast strain for production of ethanol but were unable conduct proper experiment due to practical handling error at the start of the experiment, while the WLP004 were able to produce the most biomass in this paper (Table 2), but weren’t able to produce ethanol production as much or more than the WLP500 yeast strains. For future experiment, these procedures were suggested to be repeated again in order to minimise the misconducted experimental error in this paper. Justification The paper has been reassessed for improvement following the peer-reviews from two users and self-improvement on scientific paper from website (11, 12). Based on the advice and critics that the peers provided, changes has been made in the Abstract, Introduction, Results and Discussion section. The critics from the peers are mostly on grammatical error and construction, inconsistent of past and present tense, unclear and misunderstanding in Introduction, Results and Discussion section, and the begin with a brief background on the article in the Abstract section rather than to begin with aim of the article. The Abstract has been improved by adding a general introduction and background on fermentation. The phrase and one tense per paragraph were fixed with clarity. The introduction, results and discussion sections has been improved. The sentence from previous introduction, “Yeast was traced back 3,000 yeast that the Egyptians and Babylonians utilised the fermentation of wild yeast to alveolate the bread (1) and was later then discovered by Louis Pasteur in 1845..” has been improved to “Yeast can be traced back 3,000 years where the Egyptians and Babylonians utilised the fermentation of wild yeast to alveolate the bread and is then later discovered by Louis Pasteur in 1845…” and the citation and in-text citation has been rearranged and placed at the end of each paragraph. The misunderstanding and misinterpretation in the results section has been revised to improve the clarity and the flow for the readers. The discussion section has been revised by removing unnecessary long words and maintaining the information precise and succinct. Other critics from both peers such as vague title of the paper and usage of passive voice are irrelevant for this paper for it is part of the assignment criteria. References 1. JoVE. 2014. An Introduction to Saccharomyces cerevisiae. The Journal of Visualized Experiments. 2. COFALEC. 2014. Without yeast and fermentation, no bread and no beer! COFALEC The E.U. Yeast Industry. 3. Pastor RG, Torrado RP, Garre E, Matallana E. 2011. Recent Advances in Yeast Biomass Production. Chp 11.doi:10.5772/19458. (Article.) 4. Zaragoza S. 2014. University of Texas Austin Engineer Converts Yeast Cells into ‘Sweet Crude’ Biofuel. The Univeristy of Texas at Austin. 5. Krieger C. 2008. Saccharomyces cerevisiae Pathway: superpathway of glucose fermentation. YeastCyc Biochemical Pathways. 6. Leskovac V, Trivić S, Pericin D. 2002. The three zinc-containing alcohol dehydrogenases from baker’s yeast, Saccharomyces cerevisiae. FEMS Yeast Research. 2(4): p.481-94. 7. Portela P, Moreno S. 2006. Glucose-dependent activation of protein kinase A activity in Saccharomyces cerevisiae and phosphorylation of its TPK1 catalytic subunit. Cellular Signalling. 18(7): p.1072-86. 8. Kellermann E, Seeboth PG, Hollenberg CP. 1986. Analysis of the primary structure and promoter function of a pyruvate decarboxylase gene (PDC1) from Sacchomyces cerevisiae. Nucleic Acids Research. 14(22): p.8963-77. 9. Vieira ED, Andrietta MGC, Andrietta SR. 2013. Yeast biomass production: a new approach in glucose-limited feeding strategy. Brazilian Journal of Microbiology. 44(2): p.551-8. 10. Sprague GF, Jr. 1977. Isolation and characterization of a Saccharomyces cerevisiae mutant deficient in pyruvate kinase activity. Journal of Bacteriology. 130(1): p.23241. 11. On-Line Resources Home. 2011. The Structure, Format, Content, and Style of a Journal-Style Scientific Paper. p1-17. 12. Anonymous. Twenty-One Suggestions for Writing Good Scientific Paper: Notes on Writing Papers and Theses. <http://course1.winona.edu/mdelong/ecolab/21%20suggestions.html>