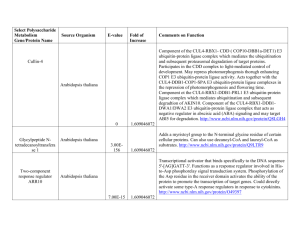
nph12963-sup-0002-TableS1-S5
advertisement
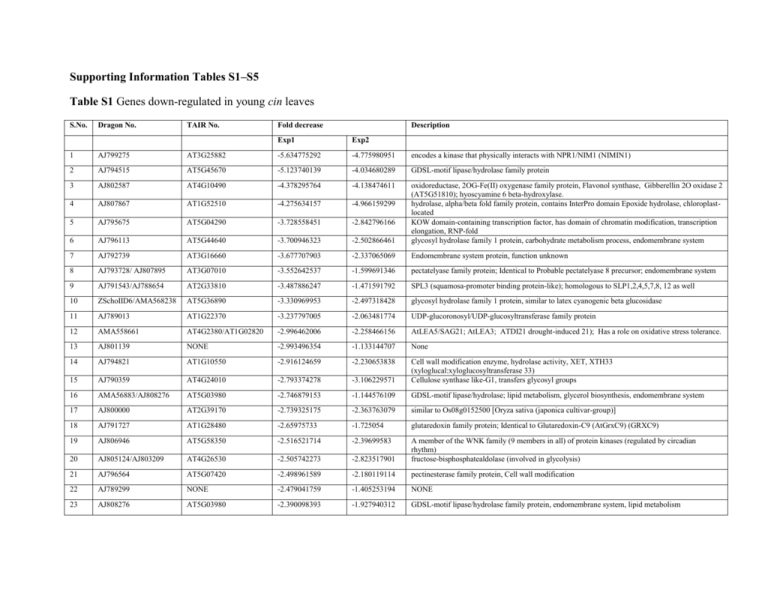
Supporting Information Tables S1–S5 Table S1 Genes down-regulated in young cin leaves S.No. Dragon No. TAIR No. Fold decrease Description Exp1 Exp2 1 AJ799275 AT3G25882 -5.634775292 -4.775980951 encodes a kinase that physically interacts with NPR1/NIM1 (NIMIN1) 2 AJ794515 AT5G45670 -5.123740139 -4.034680289 GDSL-motif lipase/hydrolase family protein 3 AJ802587 AT4G10490 -4.378295764 -4.138474611 4 AJ807867 AT1G52510 -4.275634157 -4.966159299 5 AJ795675 AT5G04290 -3.728558451 -2.842796166 6 AJ796113 AT5G44640 -3.700946323 -2.502866461 oxidoreductase, 2OG-Fe(II) oxygenase family protein, Flavonol synthase, Gibberellin 2O oxidase 2 (AT5G51810); hyoscyamine 6 beta-hydroxylase. hydrolase, alpha/beta fold family protein, contains InterPro domain Epoxide hydrolase, chloroplastlocated KOW domain-containing transcription factor, has domain of chromatin modification, transcription elongation, RNP-fold glycosyl hydrolase family 1 protein, carbohydrate metabolism process, endomembrane system 7 AJ792739 AT3G16660 -3.677707903 -2.337065069 Endomembrane system protein, function unknown 8 AJ793728/ AJ807895 AT3G07010 -3.552642537 -1.599691346 pectatelyase family protein; Identical to Probable pectatelyase 8 precursor; endomembrane system 9 AJ791543/AJ788654 AT2G33810 -3.487886247 -1.471591792 SPL3 (squamosa-promoter binding protein-like); homologous to SLP1,2,4,5,7,8, 12 as well 10 ZSchoIID6/AMA568238 AT5G36890 -3.330969953 -2.497318428 glycosyl hydrolase family 1 protein, similar to latex cyanogenic beta glucosidase 11 AJ789013 AT1G22370 -3.237797005 -2.063481774 UDP-glucoronosyl/UDP-glucosyltransferase family protein 12 AMA558661 AT4G2380/AT1G02820 -2.996462006 -2.258466156 AtLEA5/SAG21; AtLEA3; ATDI21 drought-induced 21); Has a role on oxidative stress tolerance. 13 AJ801139 NONE -2.993496354 -1.133144707 None 14 AJ794821 AT1G10550 -2.916124659 -2.230653838 15 AJ790359 AT4G24010 -2.793374278 -3.106229571 Cell wall modification enzyme, hydrolase activity, XET, XTH33 (xyloglucal:xyloglucosyltransferase 33) Cellulose synthase like-G1, transfers glycosyl groups 16 AMA56883/AJ808276 AT5G03980 -2.746879153 -1.144576109 GDSL-motif lipase/hydrolase; lipid metabolism, glycerol biosynthesis, endomembrane system 17 AJ800000 AT2G39170 -2.739325175 -2.363763079 similar to Os08g0152500 [Oryza sativa (japonica cultivar-group)] 18 AJ791727 AT1G28480 -2.65975733 -1.725054 glutaredoxin family protein; Identical to Glutaredoxin-C9 (AtGrxC9) (GRXC9) 19 AJ806946 AT5G58350 -2.516521714 -2.39699583 20 AJ805124/AJ803209 AT4G26530 -2.505742273 -2.823517901 A member of the WNK family (9 members in all) of protein kinases (regulated by circadian rhythm) fructose-bisphosphatealdolase (involved in glycolysis) 21 AJ796564 AT5G07420 -2.498961589 -2.180119114 pectinesterase family protein, Cell wall modification 22 AJ789299 NONE -2.479041759 -1.405253194 NONE 23 AJ808276 AT5G03980 -2.390098393 -1.927940312 GDSL-motif lipase/hydrolase family protein, endomembrane system, lipid metabolism 24 AJ794227 AT5G18200 -2.38654997 -1.83192884 UDP-glucose:hexose-1-phosphate uridylyltransferase, galactose metabolic process 25 ZSL1_3E10 AT2G44480 -2.159758671 -1.375341624 glycosyl hydrolase family 1 protein, endomembrane system 26 AJ790994 AT1G12940 -2.136511931 -1.471635804 member of High affinity nitrate transporter family, Enoyl CoA hydratase/isomerase domain 27 AJ791405 AT3G14395,AT1G30260 -2.134705736 -1.451963538 unknown protein., AGL79 (AGAMOUS-LIKE 79); similar to galactosyltransferase family protein 28 AJ805296 AT1G77380 -2.120345231 -1.633069012 Amino acid permease which transports basic amino acids 29 AMA560094 AT4G16730 -2.093684012 -5.618190269 30 AJ789906 NONE -2.018527246 -1.087776423 lyase/magnesium ion binding; similar to ATTPS03 (Arabidopsis thaliana terpene synthase 03), Terpenoidcyclase domain/protein phenyltransferase ?- domain NONE 31 AJ800487 AT1G80760 -1.919233364 -1.064354996 32 AJ802207 AT4G25810 -1.863748859 -1.797109309 33 AJ809153 AT5G36890 -1.8298288 -1.838001523 Similar to gb:D17443 major NIP6,NLM7,NOD26-like intrinsic protein from Oryza sativa, involved in transport, located in membrane xyloglucanendotransglycosylase-related protein (XTR6), cell wall protein, endomembrane system, domain- glycosyl hydrolase glycosyl hydrolase family 1 protein, carbohydrate metabolism 34 AJ802666 AT4G13440 -1.809197807 -1.913749559 calcium-binding EF hand family protein 35 AJ804987 AT4G37850 -1.787017167 -3.507310573 36 AJ807056 AT4G31500 -1.763451297 -1.748068486 37 AJ809024 NONE -1.758476915 -5.286743957 basic helix-loop-helix (bHLH) family protein, similar to symbiotic ammonium transporter; nodulin.(Has homology with transparent testa 8, enhancer of Glabra3, Jasmonate insensitive 1) ATR4,Cytochrome P450 Monooxygenase 83B1,Encodes an oxime-metabolizing enzyme in the biosynthetic pathway of glucosinolates. Is required for phytochrome signal transduction in red light. Mutation confers auxin overproduction. NONE 38 AMA568752 AT2G37170 -1.741906717 -2.010267698 39 AJ797470 AT4G24120 -1.736028147 -1.478853154 40 AMA568689 AT5G60660 -1.690593465 -2.116809007 41 AMA558368 AT1G23120 -1.6749256 -2.971862929 42 AJ790624 AT4G10040 -1.674395758 -1.495072421 major latex protein-related involved in defence response, similar to putative ripening-related protein, contains InterPro domain Bet v I allergen Cytochrome C2, electron carrier 43 AMA559968 AT1G65870 -1.667883803 -1.53595051 disease resistance-responsive family 44 AJ804859 AT2G33205 -1.667359656 -1.381482193 TMS membrane family protein 45 AJ793249 AT4G24000 -1.6607332 -1.800192115 46 AJ796956 AT3G26040 -1.658525541 -1.432074556 encodes a protein similar to cellulose synthase, homologous to CSLG1, CSLG2, CSLE1, CESA6(CELLULAR SYNTHASE 6),CESA5,CESA2,CESA9,CESA7or IRX3 etc transferase family protein 47 AJ808146 AT4G13440 -1.644063818 -1.075144652 calcium-binding EF hand family protein 48 AJ795075 AT2G22750 -1.629630531 -2.604336462 basic helix-loop-helix (bHLH) family 49 AJ801757 AT5G33370 -1.607744046 -1.03103383 GDSL-motif lipase/hydrolase a member of the plasma membrane intrinsic protein subfamily PIP2. localizes to the plasma membrane and exhibits water transport activity in Xenopus oocyte. expressed specifically in the vascular bundles and protein level increases slightly during leaf dev Member of a small family of oligopeptide transporters similar to the yellow stripe locus of maize (ZmYS1). PIP2F, plasma membrane, water channel 50 AJ797682 AT5G40370 -1.583018395 -1.627875433 glutaredoxin 51 AJ803945 AT4G32840 -1.540789996 -2.427842815 phosphofructokinase family protein 52 AJ796159 AT1G76470 -1.521700888 -2.144009665 cinnamoyl-CoA reductase 53 AMA568742 AT5G10760 -1.517014629 -1.330317982 aspartyl protease family protein; similar to chloroplast nucleoid DNA-binding protein 54 AJ790968 AT5G53486 -1.510815851 -1.11997327 unknown protein, Endomembrane system protein 55 AJ809133 AT1G22960 -1.464233996 -1.476050701 56 AJ805277 NONE -1.449435892 -3.333653219 pentatricopeptide (PPR) repeat-containing protein; contains InterPro domain Protein prenyltransferase. NONE 57 AMA558720 NONE -1.446520232 -1.124013179 NONE 58 AJ803222 AT2G36970 -1.442547991 -1.975523945 UDP-glucoronosyl/UDP-glucosyltransferase 59 AJ796467 AT5G47560 -1.440798919 -1.884246522 Encodes a tonoplast malate/fumarate transporter 60 AJ809141 AT1G42460 -1.413850861 -1.124691048 Ulp1 protease family protein, contains InterPro domain Peptidase C48 61 AMA568456 AT4G30210 -1.405376279 -4.258060314 62 AMA568943 AT2G30870 -1.399530909 -1.17710624 Encodes NADPH-cytochrome P450 reductase that catalyzes the first oxidative step of the phenylpropanoid general pathway. ERD13 (early dehydration-induced 13), ATGSTF10, glutathione transferasese 63 AJ796300 AT1G65445 -1.384430288 -1.558740272 transferase-related 64 AJ807059 AT4G24220 -1.376247734 -2.000399162 AWI31, VEP1 (vein patterning 1) 65 AMA560274 AT1G17420 -1.36912489 -1.365539661 Lipoxygenase,LOX3,Defence response, JA biosynthetic enz, response to wounding, groeth 66 AJ803085 AT1G43670 -1.355059865 -1.531501845 fructose-1,6-bisphosphatase, putative 67 AJ788079 AT1G67910 -1.33951 -1.77238 similar to unknown protein, serine rich protein related 68 AJ806443 AT3G61220 -1.339158836 -3.346235844 SDR (short chain dehydrogenase/reductase) family 69 AJ797289 AT2G45680 -1.34 -1.88 TCP family transcription factor, [TCP 9],contains InterPro domain TCP transcription factor 70 AJ797785 NONE -1.333851986 -1.016238555 NONE 71 AJ804448 AT1G32780 -1.33149651 -1.326894455 alcohol dehydrogenase 72 AJ803635 AT3G49260 -1.329955541 -1.02668888 IQ-domain 21, calmodulin binding 73 AJ809139 AT1G49620 -1.318403572 -1.108608998 KRP7/ICN6/ICK5, (6e-15) 74 AJ803180 AT2G14750 -1.309206114 -2.142657705 AKN1/ATAKN1/APK (APS kinase) (5e-73) 75 AY534686 AT4G38460 -1.307755507 -4.449999937 76 AJ806669 NONE -1.305881484 -1.784018895 GGR (GERANYLGERANYL REDUCTASE); farnesyltranstransferase; Identical to Geranylgeranyl pyrophosphate synthase-related protein, chloroplast precursor (GGR), contains InterPro domain Terpenoid synthase NONE 77 AJ808569 NONE -1.287074649 -1.580016342 NONE 78 AJ807170 AT2G33590 -1.280116874 -2.369665211 cinnamoyl-CoA reductase family 79 AJ795833 AT3G63250 -1.276042039 -1.115459499 ATHMT-2/HMT-2; homocysteine s-METHYLTRANSFERASE 80 AJ793435 NONE -1.272926117 -1.56582676 NONE 81 AJ794513 AT2G47370 -1.272271052 -1.291800754 UNKNOWN (Alcohol Dehydrogease-like domain) 82 AJ788963 AT1G43670 -1.253175608 -1.409358318 fructose-1,6-bisphosphatase, putative 83 AMA558767 AT4G21990 -1.237563617 -2.013754807 PRH-26, ATAPR3, APS Reductase 3) 84 AJ792044 AT3G53420 -1.23175926 -1.945259415 PIP2A (PLASMA MEMBRANE INTRINSIC PROTEIN 2; WATER CHANNEL) 85 AJ791944 NONE -1.211639093 -2.699957549 NONE 86 AJ804255 AT4G19840 -1.20272101 -1.569411552 ATPP2-A1 (Arabidopsis thaliana phloem protein 2-A1) 87 AJ794089 AT3G51895 -1.168308862 -1.545123359 AST12, ATST1, SULTR3;1, SULTR3 (SULFATE TRANSPORTER) 88 AJ803687 AT1G70810 -1.168217364 -1.389696048 C2-DOMAIN CONTAINING PROTEIN 89 AJ791963 AT5G04200 -1.166961873 -2.716303578 LATEX-ABUNDANT PROTEIN, AMC9: CASPASE FAMILY PROTEIN 90 AJ804255 AT4G19840 -1.159849635 -1.350141039 ATPP2-A1 (A. thaliana PHLOEM PROTEIN 2-A1) 91 AJ805825 AT2G23620 -1.152621661 -3.030836525 ESTERASE, PUTATIVE 92 AJ799455 AT2G14750 -1.152355475 -2.33915066 AKN1, APK (APS Kinase) 93 AJ797898 AT4G24120 -1.151625381 -1.796001155 YSL1 (YELLOW STRIPE LIKE 1); OLIGOPEPTIDE TRANSPORTER 94 AJ801395 AT5G62280 -1.138507007 -1.73828121 UNKNOWN 95 AJ794980 AT4G37990 -1.13576727 -4.491745151 ELI3-2 (ELICITOR-ACTIVATED GENE 3) 96 AJ790744 AT1G68570 -1.125202279 -1.285049375 PROTON-DEPENDENT OLIGOPEPTIDE TRANSPORT (POT) FAMILY PROTEIN 97 AJ789851 AT2G34070 -1.114105818 -1.263588981 UNKNOWN 98 AMA559497 NONE -1.112828381 -1.812461582 NONE 99 AJ795752 NONE -1.10523534 -1.403217634 NONE 100 AJ801897 AT5G15840 -1.100452241 -1.87204692 CONSTANS 101 AJ795364 AT5G50790 -1.059182798 -2.486413206 nodulin MtN3 family protein; senescence-associated protein (SAG29) [Arabidopsis thaliana] 102 AJ796089 AT3G61040 -1.028715191 -1.496453504 encodes a protein with cytochrome P450 domain 103 AJ799167 AT5G48810 -1.022718318 -2.475005434 Encodes a cytochrome b5 isoform 104 AMA568850 AT5G47700 -1.019373143 -2.910604143 60S acidic ribosomal protein P1 (RPP1C); InterPro domain Ribosomal protein 60S 105 AJ809130 NONE -1.017248349 -1.440018414 NONE 106 AJ790214 AT3G15353 -1.010397079 -2.820971381 metallothionein, binds to and detoxifies excess copper and other metals, limiting oxidative damage 107 AJ792815 NONE -1.010353428 -1.705618394 NONE 108 AJ807951 AT2G45550 -1.008282176 -1.762707323 member of CYP76C, CYP76C4, CYTOCHROME P450, FAMILY 76, SUBFAMILY C 109 AJ796269 AT2G45570 -1.000990519 -1.655626739 member of CYP76C Table S2 Genes up-regulated in young cin leaves S.No. Dragon No. TAIR No. Fold Increase Description Exp1 Exp2 1 AMA559965 NONE 4.632589 4.875015 NONE (Up-1) 2 AJ796073 AT3G12580 3.158397 2.240245 HSP 70 3 AJ792486 AT4G37260 3.064101 1.968502 MYB 73, regulation of transcription; DNA dependent, response to- ABA, Auxin, Ethylene, GA, JA,SA, Cadmium ion 4 AJ792505 AT5G61600 3.002922 1.379206 Ethylene response factor (AP2 class) 5 AJ787178 AT1G70850 2.963041 1.956739 Bet v I allergen family protein; Identical to MLP-like protein 34 (MLP34) 6 AJ790801 AT2G38470 2.911741 1.263767 7 AJ790792 AT4G23140 2.721937 2.818696 WRKY DNA-BINDING PROTEIN 33,[ regulation of transcription, DNA-dependent, defense response to bacterium, defense response to fungus] Cysteine-rich receptor like kinase 8 AJ795483 AT5G35200 2.536321 1.685408 epsin N-terminal homology (ENTH) domain-containing protein; Identical to Putative clathrin assembly protein At5g35200 9 AJ800324 AT5G39670 2.504609 1.579522 calcium-binding EF hand family protein 10 AJ791652 AT1G62510 2.369444 3.619665 Protease inhibitor/seed storage/lipid transfer protein (LTP) family protein; similar to ELP (EXTENSIN-LIKE PROTEIN) 11 AJ798250 NONE 2.331414 2.084555 NONE 12 AJ805666 AT3G23980 2.196373 2.822087 13 AMA559197 AT5G03980 2.187913 2.393685 similar to CIP1, similar to heavy meromyosin-like protein; similar to Os01g0911800 [Oryza sativa (japonica cultivar-group)] (GB:NP_001045163.1); contains InterPro domain Spectrin repeat; (InterPro:IPR002017) GDSL-motif lipase/hydrolase family protein; similar to lipase 14 AJ806613 AT5G12960 2.183778 2.239038 similar to unknown protein 15 AMA558863 AT1G59560 2.166284 2.550161 ZCF61; protein binding / zinc ion binding; similar to zinc finger family protein 16 AJ803089 AT4G25470 2.13868 1.719742 17 AJ791638 AT2G44670 2.138231 1.242138 Encodes a member of the DREB subfamily A-1 of ERF/AP2 transcription factor family (CBF2). The protein contains one AP2 domain. senescence-associated protein-related 18 AJ806624 AT1G20440 2.044922 2.260188 19 AJ796336 AT1G32640 1.980855 1.975367 20 AMA558864 AT1G69120 1.973258 2.406864 21 AJ803092 AT1G30040 1.711272 1.163428 Belongs to the dehydrin protein family, which contains highly conserved stretches of 7-17 residues that are repetitively scattered in their sequences, the K-, S-, Y- and lysine rich segments. Encodes a 68 kD MYC-related transcriptional activator with a typical DNA binding domain of a basic region helix-loop-helix leucine zipper motif. Floral homeotic gene encoding a MADS domain protein homologous to SRF transcription factors. Specifies floral meristem and sepal identity. Required for the transcriptional activation of AGAMOUS. Interacts with LEAFY Encodes a gibberellin 2-oxidase. AtGA2OX2 expression is responsive to cytokinin and KNOX activities 22 AJ794180 NONE 1.709118 2.429822 NONE 23 AJ790183 NONE 1.602345 1.492777 NONE 24 AMA558584 NONE 1.587897 1.477305 NONE 25 AJ790343 AT4G37990 1.536932 1.637402 26 AJ804131 AT4G29190 1.53086 1.034481 Encodes an aromatic alcohol:NADP+ oxidoreductase whose mRNA levels are increased in response to treatment with a variety of phytopathogenic bacteria. zinc finger (CCCH-type) family protein; similar to zinc finger (CCCH-type) family protein 27 AJ789493 AT4G37970 1.516412 1.654755 mannitol dehydrogenase, putative; similar to ELI3-2, similar to sinapyl alcohol dehydrogenase-like protein. 28 AJ793815 AT2G19810 1.467851 1.46861 zinc finger (CCCH-type) family protein; similar to zinc finger (CCCH-type) family protein 29 AMA559563 AT3G02000 1.453428 1.813553 30 AJ803437 AT1G50600 1.442989 1.089546 Roxy1 encodes a glutaredoxin belonging to a subgroup specific to higher plants. It is required for proper petal initiation and organogenesis. Encodes a scarecrow-like protein (SCL5). Member of GRAS gene family 31 AJ792444 NONE 1.440153 1.194919 NONE 32 AMA559761 AT4G39330 1.42418 1.110323 mannitol dehydrogenase, putative; Identical to Probable mannitol dehydrogenase 33 AJ789267 AT2G46490 1.381744 1.492912 Similar to unknown protein 34 AJ809148 NONE 1.379153 1.150418 NONE 35 AJ809113 AT4G29780 1.378719 2.257841 Similar to unknown protein 36 AJ800329 AT2G19810 1.352487 1.584558 zinc finger (CCCH-type) family protein 37 AJ805745 AT5G03980 1.343361 1.840284 GDSL-motif lipase/hydrolase family protein; similar to lipase 38 AJ806658 AT1G14820 1.331185 1.201106 SEC14 cytosolic factor family protein / phosphoglyceride transfer family protein; similar to SEC14 cytosolic factor 39 AJ798394 AT5G10100 1.295085 1.078101 Similar to trehalose-6-phosphate phosphatase 40 AJ808571 AT5G45390 1.291333 1.862421 One of several nuclear-encoded ClpPs (caseinolytic protease). Contains a highly conserved catalytic triad of Ser-type proteases 41 AJ800479 AT4G14270 1.267793 1.197725 Protein containing PAM2 motif which mediates interaction with the PABC domain of polyadenyl binding proteins. 42 AJ792118 AT3G55500 1.265621 1.095421 Expansin-like protein. Involved in the formation of nematode-induced syncytia in roots of Arabidopsis thaliana 43 AJ798626 AT5G05960 1.260809 1.541136 protease inhibitor/seed storage/lipid transfer protein (LTP) family protein 44 AMA560140 AT4G27280 1.251728 2.24731 calcium-binding EF hand family protein; similar to PBP1 (PINOID-BINDING PROTEIN 1) 45 AJ809029 AT1G19020 1.248985 1.100898 similar to unknown protein 46 AJ804190 AT4G25470 1.206219 2.099331 47 AJ790933 AT5G14070 1.181438 1.298345 Encodes a member of the DREB subfamily A-1 of ERF/AP2 transcription factor family (CBF2). The protein contains one AP2 domain glutaredoxin family protein; Identical to Glutaredoxin-C8 (AtGrxC8) (GRXC8) 48 AJ796270 AT2G38185 1.174352 1.004719 zinc finger (C3HC4-type RING finger) family protein; similar to unknown protein 49 AJ809015 AT3G55840 1.165042 1.386389 similar to unknown protein, similar to putative Hs1pro-1-like receptor, contains InterPro domain Hs1pro-1 50 AJ802811 AT3G54720 1.156127 1.04152 Encodes glutamate carboxypeptidase 51 AJ798949 NONE 1.145477 1.184797 NONE 52 AJ808942 AT3G10410 1.134078 1.150721 SCPL49 (serine carboxypeptidase-like 49); serine carboxypeptidase; Identical to Serine carboxypeptidase precursor 53 AJ802728 AT5G10100 1.131951 1.133392 trehalose-6-phosphate phosphatase 54 AJ794642 AT3G23930 1.117225 1.117225 similar to unknown protein 55 AMA560140 AT4G27280 1.091185 2.423884 calcium-binding EF hand family protein; similar to PBP1 (PINOID-BINDING PROTEIN 1) 56 AJ789950 AT4G36500 1.067795 1.251367 similar to unknown protein 57 AJ798414 AT1G74660 1.052609 1.033828 58 AJ790546 AT5G67090 1.036161 1.070495 Constitutive overexpression of MIF1 caused dramatic developmental defects, seedlings were non-responsive to GA for cell elongation [ZF-HD homeobox protein Cys/His-rich dimerisation ] similar to subtilase family protein 59 AMA560239 AT4G25760 1.033272 1.540935 similar to unknown protein Table S3 PCR primers and oligos used in the study Primer Number Primer Sequence (5'-3') Antirrhinum EST (Closest Arabidopsis Homolog) P01597 Forward (FP)/ Reverse (RP) Primer FP AGGTTGGGATTGCCTGGAGCTGA AJ795150 (IAA1) P01598 RP TGTGCCTTTGGAGCTGGAGCA AJ795150 (IAA1) P01599 FP GGAACACGACCTCCTCCGGC AJ798118 (IAA7) P01600 RP GAGCCCCGTCAACTGCCACC AJ798118 (IAA7) P01601 FP CCGTCGGATGTGAGGTCGTGC AJ801045 (TGSAUR21) P01602 RP GCAACCGCACACGATCACCG AJ801045 (TGSAUR21) P01603 FP GTCCGTACCCCGCGGACCAA AJ803900 (ARG10) P01604 RP TCCCCAGTACAGCTTCACGCCA AJ803900 (ARG10) P01605 FP AGCAGATGCCAGAGGATTGGGC AJ790979 (IAA3) P01606 RP AGACTCTTCGAGAACCCAAGCTGT AJ790979 (IAA3) P01607 FP AGGGCTACCGGGAACTGATAACGA AJ799874 (IAA3) P01608 RP TCGAACCGGAGGCCATCCAACT AJ799874 (IAA3) P01609 FP TGCTGCTGCTAAGAACAACAAGCG AJ804892 (IAA3) P01610 RP CCGGTAAGACCTCACTGGTGGC AJ804892 (IAA3) P01611 FP TCACCAGGTGTCCGTGGGTTGT AJ807692 (HK4) P01612 RP ACTGCACGTCAGCCCCAAGC AJ807692 (HK4) P01613 FP TGAGGTCCATGTTCTCGCGGTT AJ804107 (ARR3) P01614 RP AGCCCTCCTCCCACTGTCCA AJ804107 (ARR3) P01615 FP ACACCATCACCGTCGCCCCT AJ787388 AND AJ801951 (ARR15) P01616 RP GGCAGCCCCATCAGTTTGGAGC AJ787388 AND AJ801951 (ARR15) P01617 FP TGGAGGCGAGGATCTTGCATGG AJ806940 (ARR6) P01066 RP GCTTCAGGTGGACCCTTGGCC AJ806940 (ARR6) P01067 FP CCACTTCTCCCAGCTCGACCAAC AJ799891 (APRR7) P01618 RP CGCTTCCCAATTACTCTGCGCCA AJ799891 (APRR7) P01619 FP GGACGGCGGTTTGGGACTGT AJ792505 (Ethylene response factor-AP2 class) P01620 RP CCCCAGACTATTACCGTCAGGTGC AJ792505 (Ethylene response factor-AP2 class) P01621 FP AGTCCGGGTTCTCCCCTGGA AJ806643 (LOX2) P01622 RP CCGCCACAGCCACTCCCCTA AJ806643 (LOX2) P01072 FP GTACTTCGGCGTCGCCATGCC AMA560274 (LOX3) P01073 RP GCTTAGCCGCACCAACGAGTGTC AMA560274 (LOX3) P01623 FP TCGACGGGAACTGACGTTCCAA AJ798778 (putative auxin-inducible SAUR gene) P01624 RP CACCCATTGGATGGTTGAACCCAAA AJ798778 (putative auxin-inducible SAUR gene) P01625 FP GGCGGATAAGCTAGGCGGGGA AMA560140 (similar to PBP1 - PINOID-BINDING PROTEIN 1) P01626 RP CCCCTCCTGTTGAAGTGCCTCT AMA560140 (similar to PBP1 - PINOID-BINDING PROTEIN 1) PO1660 FP AGCTGAGGTTGGGATTGCCTGGA AJ795082 (IAA1) PO1661 RP TGCCCTTGGAGCTGGAGCAGA AJ795082 (IAA1) PO1662 FP GCCTTCTTCACCTCCTGGCCG AJ795336 (IAA12/11) PO1663 RP GGCTGGCGATTTTGTATGGTTTGCC AJ795336 (IAA12/11) PO1664 FP TCACGCCATGTGCAACTCCGT AJ791402 (IAA4/5) PO1665 RP GCGCAACAGATCGTCGAGACCAC AJ791402 (IAA4/5) PO1666 FP AGCTGAGATTGGGTTTGCCTGGT AY062215 (IAA4 AUX/IAA1) PO1667 RP TGTGCCTTTGGAGCTGGTGCT AY062215 (IAA4 AUX/IAA1) PO1668 FP AGCTTTGCCCGGCGATGAGG AJ568789 (IAA16) PO1669 RP GGCATTGGGGGCCATCCCAC AJ568789 (IAA16) PO1670 FP TGAGCTTAGATTGGGTTTGCCTGGT AJ802114 (IAA4) PO1671 RP TGGAGCTGGTGCTATTGATTCCTCT AJ802114 (IAA4) PO1672 FP GTGGTGGGATGGCCTCCGGT AJ793214 (IAA16) PO1673 RP GCGCTCCGTCCATGCTCACC AJ793214 (IAA16) PO1674 FP TGCAAGCGCCTTCGCATCAT AJ559808 (IAA16) PO1675 RP TGCTCCATCAAACCAAGTTGCACA AJ559808 (IAA16) PO1676 FP GTGACTGGATGCTCGTGGGGG AJ791366 (IAA16) PO1677 RP TGCACTTCTCCACTGCTCTTGGT AJ791366 (IAA16) PO1678 FP TCCCCTGCCAAGGCCCAAGT AJ806977 (IAA16) PO1679 RP CGGTGCCCCATCAACGCTCA AJ806977 (IAA16) PO1680 FP TCTGTCGGAAAGTGGAACTATGGC AJ789312 (ARR15) PO1681 RP CAAATCCAACAGAACCCCTTTCTCC AJ789312 (ARR15) PO1682 FP ACCAGGAACCCCCAATGCAGC AJ558353 (APRR7) PO1683 RP GGGGCATCAGCCGCCAACTT AJ558353 (APRR7) PO1684 FP GGCACGAGTGAATGCACACAGTTT AJ808985 (AHP4) PO1685 RP TCGAGCTTCCTTTTCAGGACTGC AJ808985 (AHP4) P00330 FP AGGCCGATTGTGCTGTTCTG Antirrhinum EF1 P00331 RP GAAAGCAAGCAATGCATGCTC Antirrhinum EF1 P00332 FP CCACATTCGCAGTCCCATC CINCINNATA P00333 RP GAAACGACTGAGCTCCGCAG CINCINNATA RT-PCR and cloning the fragments into pGEM T - Easy for in-vitro transcription P00939 FP TGACGATACAATCTTGTCCAAGGG AJ807692 (HK4) P00940 RP TTGGTATGTGCCACTCGGTAC AJ807692 (HK4) P00265 FP TGGCAAGTGTTATGGCTCATGGAAC AJ790979 (IAA3) P00266 RP TCAAACAGAACAGCCCAATCCTCTG AJ790979 (IAA3) Primers used for ChIP Primer Number Primer Sequence (5'-3') Gene/Binding Site PO1783 FP GAGTACGCATATTTAAAGGTCAACTCGC AmHK4 BS1 PO1784 RP GTACTGATTCAGTAAACTGCTTGGGG AmHK4 BS1 PO1785 FP CAGTTTATTCTTATGCGCTAATGTTGATG AmHK4 BS2 PO1786 RP CCATCCATTGACCCTTAGTTAACTATC AmHK4 BS2 PO1787 FP ACGGGAGGGCAGAGCCTGAT AmHK4 BS3 PO1788 RP AAGTGAGGGAGGGAGGGCGA AmHK4 BS3 PO1789 FP CCATTTCTGTTTCTCTCTCACAAGTCC AmHK4 BS4 PO1790 RP CCTGCATTTGGGTCCTCAGAATAAG AmHK4 BS4 PO1791 FP CAGGATACGTTTGCTGAATACACCGC AmHK4 BS5 PO1792 RP TGACAGGAGCATACTCATCCCTTGC AmHK4 BS5 PO1793 FP CATGGCTTTCCTAGTCTCTTCTCTCC AmHK4 BS6 PO1794 RP GAGAGTGATCGTGTGTATCTTTCTTCAGC AmHK4 BS6 PO1795 FP CATCACCGGTCGGGTAACATAACTTTC AmHK4 BS7 PO1796 RP CGGGAAGAACTACATTAGGTGGGC AmHK4 BS7 PO1797 FP GCCGATGTGGCTAAATCTCAGGTTC AmHK4 BS8 PO1798 RP GCAGTGCCAACATCCCTAAAAGCAC AmHK4 BS8 PO1799 FP GAGAGGAAAGTGTCGGAAATGATTTATG AmHK4 BS9 PO1800 RP GAGAGCAATTTGTGAACACCTGCAATA AmHK4 BS9 PO1261B FP GGAAACCAAGACTCTGATTCATAACC AmIAA3 BS1 PO1262 FP GGTGCCTCCCATGTGGAGGAG AmIAA3 BS1 PO1263 RP GGAATCAGGTGTGTCCGATTTGG AmIAA3 BS1 PO1264 FP GTTCAAGCTCACCATAGGTAGG AmIAA3 BS2 PO1265 RP GTAGCCTTCTCTCTCTGAGTATTC AmIAA3 BS2 PO1266 FP GAGAAGAAGATGCTGTATCTGAGG AmIAA3 BS3 PO1267 RP ACAGGATTACAAAACCGCCAAGGC AmIAA3 BS3 Primers used for EMSA PO1801 FP TTTTTATTGGCCCTGCAACT AmHK4 BS1 PO1802 RP AGTTGCAGGGCCAATAAAAA AmHK4 BS1 PO1803 FP ATATTAATGGACCAATATGA AmHK4 BS2 PO1804 RP TCATATTGGTCCATTAATAT AmHK4 BS2 PO1805 FP GTGGAAGGTGGACCTTTTGGT AmHK4 BS3 PO1806 RP ACCAAAAGGTCCACCTTCCAC AmHK4 BS3 PO1807 FP TGCAACAGGACCATGCTTTA AmHK4 BS4 PO1808 RP TAAAGCATGGTCCTGTTGCA AmHK4 BS4 PO1809 FP AGCATGGGTGGACCATTCGGA AmHK4 BS5 PO1810 RP TCCGAATGGTCCACCCATGCT AmHK4 BS5 PO1811 FP CTTGGTTTGGACCTGCTGAA AmHK4 BS6 PO1812 RP TTCAGCAGGTCCAAACCAAG AmHK4 BS6 PO1813 FP CAAGTAAGGGCCAATTCCAT AmHK4 BS7 PO1814 RP ATGGAATTGGCCCTTACTTG AmHK4 BS7 PO1815 FP TGTGTAAGGACCATTTTTTT AmHK4 BS8 PO1816 RP AAAAAAATGGTCCTTACACA AmHK4 BS8 PO1817 FP CATGCTGGTGGGCCTCTTAGC AmHK4 BS9 PO1818 RP GCTAAGAGGCCCACCAGCATG AmHK4 BS9 PO1256 FP ACTACTAGGCCCACATGGTCA AmIAA3 BS1 PO1257 RP TGACCATGTGGGCCTAGTAGT AmIAA3 BS1 PO1258 FP TGTTGAATGGTCCTTGGATA AmIAA3 BS2 PO1259 RP TATCCAAGGACCATTCAACA AmIAA3 BS2 PO1260 FP TTTTATCTGGACCTCATCTG AmIAA3 BS3 PO1261A RP CAGATGAGGTCCAGATAAAA AmIAA3 BS3 Primers used for SELEX G704 FP GGAAACAGCTATGACCA TG(N)18ACTGGCCGTCGTTTTAC RP GTAAAACGACGGCCAGT Primers used for cloning in protoplast assays 5742 FP CCGCTCGAGCATGGGAGGAGCAGGAGGA CIN ORF 5743 RP CCGCTCGAGTCAATGGCGAGATTCGGAG CIN ORF P02118 FP TAGGATCCCGTCCGAAGGCTGAAACCACGC AmIAA3 promoter P02119 RP TACCATGGGGTTGGAGTCTTTGGTGTTTTGGG AmIAA3 promoter P02225 FP ATTCTGCAGCTCTGATTCATAACCCCAACCCCTC AmIAA3 promoter P02226 FP AAACTGCAGCACCAACCACCCATGTAGACCCTC AmIAA3 promoter P02227 RP AAACCATGGCGTATCCTGTATTACCCACGGAAGC AmIAA3 promoter P02183 FP TTCTGCAGCTAAGGGTCAATGGATGGAAGGG AmHK4 promoter P02184 FP TTCTGCAGCCTCCCTTCCATCCATTGACCC AmHK4 promoter P02184 RP AAACCATGGAAAATATGACAGGAGCATACTCATCC AmHK4 promoter P02123 RP TACCATGGGCAAGCTTTGTGAGTGACTGAACAC AmHK4 promoter Table S4 Genes involved in auxin and cytokinin signaling differentially expressed in cin leaf Auxin signaling pathway Sl. No. Dragon ID 1. 2. 3. 4. 5. 6. 7. 8. 9. 10. 11. 12. 13. 14. 15. 16. 17. 18. 19. 20. 21. 22. 23. 24. 25. AJ799874 AJ804892 A_84_P21812 AJ790979 AJ798118 AJ795082 AJ795336 AJ806069 AJ791402 AY062215 AJ568789 AJ802114 AJ793214 AJ559808 AJ791366 AJ806977 AJ801045 AJ559301 AJ793092 AJ798778 AJ789007 AJ803535 AJ792146 ----- TAIR ID Closest Homologue in Arabidopsis Young leaf AT1G04240.1 AT1G04240.1 AT1G04240.1 AT4G14550.1 AT1G04240.1 AT4G28640.2 AT1G04550.2 AT5G43700.1 AT5G43700.1 AT3G04730.1 AT5G43700.1 AT3G04730.1 AT3G04730.1 AT3G04730.1 AT3G04730.1 AT5G20820.1 AT3G12830.1 AT4G38840.1 AT4G38840.1 AT4G38840.1 AT3G60690.1 AT4G38840.1 At4g34770 At4g34810 IAA3/SHY2 IAA3/SHY2 IAA11 homolog IAA3/SHY2 IAA14/ SLR IAA3/SHY2 IAA11 IAA12/BDL IAA4/ATAUX2-11 IAA4/ATAUX2-11 IAA16 IAA4/ATAUX2-11 IAA16 IAA16 IAA16 IAA16 SAUR-like auxin-responsive protein SAUR-like auxin-responsive protein SAUR-like auxin-responsive protein SAUR-like auxin-responsive protein SAUR-like auxin-responsive protein SAUR-like auxin-responsive protein SAUR-like auxin-responsive protein SAUR-like auxin-responsive protein SAUR-like auxin-responsive protein Fold change Mature Base/Tip leaf +13.5 -2.5 +28.3 -1.7 -2.2 -5.6 +3.3 +2.1 +2.2 +3.9 +8.4 +4.9 4.4 +3.7 +2.3 -3 +3.4 +2.3 -2.5 -2.2 -2.1 +4.1 -23 -4.6 2.9 +60.9 -2.7 -4.1 -2.4 2.6 Cytokinin signaling pathway Sl. No. Dragon ID TAIR ID Closest Homologue in Arabidopsis Young leaf 1. 2. 3. 4. 5. 6. 7. 8. 9. AJ807692 AJ787388 AJ789312 AJ804107 AJ799891 AJ558353 AJ808985 AJ808120 AJ559786 AT1G27320 AT1G74890 AT1G74890 AT1G59940 AT5G02810 AT5G02810 AT3G16360 AT5G53290 AT4G23750 AHK4 homologue ARR15 ARR15 ARR3 APRR7 APRR7 AHP4 CRF3 CRF2 Fold change Mature Base/Tip leaf -38.4 -2.6 -3.3 -3.6 -4.3 -3.5 +4.7 +28.3 -2.3 -2.1 -2.4 -9.3 +9.3 +7.6 Table S5 Accession number of genes used for sequence analysis and phylogeny Gene Species Accession Sas SHK300 Striga asiatica ABG35782 Sas SHK279 Striga asiatica ABG35783 Ptr CR1A Populus trichocarpa ACE63264 Vvi HK4-like Vitis vinifera XP_002285117 Rco HK-like Ricinus communis XP_002527541 Ptr CR1B Populus trichocarpa ERP57582 Bpe CR1-like Betula pendula ACE63259 Gma HK4-like Glycine max XP_003530935 Zma HK3-like Zea mays BAE80688 Osa HK1-like Oryza sativa ABF98563 Bpe HK2-like Betula pendula ACE63260 Cro CKR1 Catharanthus roseus AAM14700 Vvi HK2-like Vitis vinifera XP_002269977 Sly HK2-like Solanum lycopersicum XP_004243558 Csa HK2-like Cucumis sativus XP_004140009 Gma HK2-like Glycine max XP_003545073 TAIR ID Gma HK4-like Glycine max XP_003531201 Msa HK1 Medicago sativa ABJ74169 Lja HK2 Lotus japonicus ABI48270 Lal HK1 Lupinus albus ABJ74170 Zma HK3 Zea mays BAD01586 Mtr HK Medicago truncatula XP_003617960 Cma HK Cucurbita maxima CAF31355 Sbi HK-like Sorghum bicolor XP_002466563 Hvu HK-like BAJ89805 Osa HK2-like Hordeum vulgare subsp. vulgare Oryza sativa Smo CR1 Selaginella moellendorffii XP_002967800 Phy HK-like Petunia x hybrida BAL43559 Ppt CRE1 Physcomitrella patens ABD49494 Ppt CRE2 Physcomitrella patens ABD49495 Ppt HK4-like Physcomitrella patens XP_001779177 At HK1 Arabidopsis thaliana 816291 AT2G17820 At HK2 Arabidopsis thaliana 833552 AT5G35750 At HK3 Arabidopsis thaliana 839621 AT1G27320 BAD88305 At HK4, CRE1 Arabidopsis thaliana 814714 AT2G01830 IAA3, SHY2 Arabidopsis thaliana 839570 AT1G04240 IAA6 Arabidopsis thaliana 841717 AT1G52830 IAA8 Arabidopsis thaliana 816798 AT2G22670 IAA9 Arabidopsis thaliana 836693 AT5G65670 IAA10 Arabidopsis thaliana 839290 AT1G04100 IAA13 Arabidopsis thaliana 817894 AT2G33310 IAA16 Arabidopsis thaliana 819633 AT3G04730 ARF11, ARF19, IAA22 Arabidopsis thaliana 838505 AT1G19220 IAA26, PAP1 Arabidopsis thaliana 820898 AT3G16500 IAA27, PAP2 Arabidopsis thaliana 829029 AT4G29080 IAA29 Arabidopsis thaliana 829361 AT4G32280 IAA31 Arabidopsis thaliana 821026 AT3G17600 IAA33 Arabidopsis thaliana 835848 AT5G57420 AXR5, IAA1 Arabidopsis thaliana 827103 AT4G14560 IAA2 Arabidopsis thaliana 821877 AT3G23030 IAA5, AUX2-27 Arabidopsis thaliana 838128 AT1G15580 IAA6, SHY1 Arabidopsis thaliana 841717 AT1G52830 IAA7 Arabidopsis thaliana 821879 AT3G23050 IAA11 Arabidopsis thaliana 828982 AT4G28640 IAA12, BDL Arabidopsis thaliana 839495 AT1G04550 IAA14 Arabidopsis thaliana 827102 AT4G14550 IAA15 Arabidopsis thaliana 844379 AT1G80390 IAA18 Arabidopsis thaliana 841623 AT1G51950 IAA20 Arabidopsis thaliana 819313 AT2G46990 IAA21, IAA23, IAA25, ARF7 IAA30 Arabidopsis thaliana 832196 AT5G20730 Arabidopsis thaliana 825383 AT3G62100 IAA32 Arabidopsis thaliana 814648 AT2G01200 IAA34 Arabidopsis thaliana 838070 AT1G15050