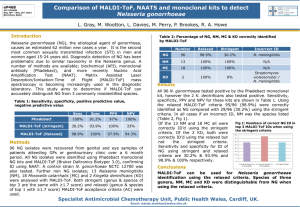
Graph 1. Time to Identification, MALDI
advertisement

TITLE: USE OF MATRIX ASSISTED LASER DESORPTION IONIZATION –TIME OF FLIGHT MASS SPECTROMETRY IN A PEDIATRIC CLINICAL LABORATORY FOR IDENTIFICATION OF BACTERIA COMMONLY ISOLATED FROM CYSTIC FIBROSIS PATIENTS RUNNING TITLE: MALDI-TOF MS FOR BACTERIAL IDENTIFICATION IN CF Ankita P. Desai 1, Theresa Stanley2, Maria Atuan2, Jonelle McKey2, John J. LiPuma4, Beverly B. Rogers2,3, Robert C. Jerris2,3 1 Department of Pediatric Infectious Diseases, Emory University School of Medicine, 2015 Uppergate Drive, Atlanta, Georgia 30322 2 Department of Pathology, Children’s Healthcare of Atlanta , 1405 Clifton Road NE, Atlanta, Georgia 30322 3 Department of Pathology, Emory University School of Medicine, 2015 Uppergate Drive, Atlanta, Georgia 30322 4 Department of Pediatrics, University of Michigan, 1415 Washington Heights, Ann Arbor, Michigan 48109 Corresponding Author: Ankita Patel Desai, MD Pediatric Infectious Diseases Fellow Emory University School of Medicine 2015 Uppergate Drive Atlanta, GA 30322 Phone: 404 727 5642 Fax: 404 727 9223 E-mail: ankita.desai@emory.edu Part of this work was presented at the American Society for Microbiology 110 th General Meeting in May of 2010 in San Diego, California. This work was supported by a grant from Bruker Daltonic, Billerica, MA, USA. Use of Matrix Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry in a Pediatric Clinical Laboratory for Identification of Bacteria Commonly Isolated from Cystic Fibrosis Patients Ankita P. Desai1, Theresa Stanley2, Maria Atuan2, Jonelle McKey2, John J. LiPuma4, Beverly B. Rogers2,3, Robert C. Jerris2,3 1 Department of Pediatric Infectious Diseases, Emory University School of Medicine, Atlanta, Georgia 2 Department of Pathology, Children’s Healthcare of Atlanta, Atlanta, Georgia 3 Department of Pathology, Emory University School of Medicine, Atlanta, Georgia 4 Department of Pediatrics, University of Michigan, Ann Arbor, Michigan ABSTRACT: Background: Matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) has been described as a rapid, accurate method for bacterial identification. We investigated the ability of the technique, using the unamended company database to identify bacteria commonly isolated in cystic fibrosis (CF) patients. Methods: We compared organisms commonly isolated from cystic fibrosis (CF) patients identified by MALDI-TOF MS (Bruker Daltonic, Billerica, MD) to conventional phenotypic and genotypic analysis. For MALDI-TOF MS, the direct colony technique was used routinely with one extraction procedure performed on a mucoid Pseudomonas aeruginosa. For 24 unique cystic fibrosis specimens, workload comparison and time to identification was assessed. Results: Of 464 tested isolates, conventional (phenotypic and genotypic) identification compared to MALDI-TOF MS showed complete genus, species agreement in 92%, with genus agreement in 98%. This included 29 isolates among the Burkholderia cepacia complex. All 29 were correctly identified to the genus level and 24 of these were speciated. Time to identification with 48 bacterial isolates from 24 CF patients revealed identification of 85% of isolates by MALDI-TOF MS at 48 hours of incubation, compared to only 34% with conventional methods. Conclusions: Using the unamended database supplied with the system, MALDI-TOF MS provides rapid and reliable identification of bacteria isolated from CF specimens. Time to identification studies concluded the use of same day, same method for organism identification will decrease time to result and optimize microbiology workflow. INTRODUCTION: Progress in rapid diagnostics has seen an unparalleled growth with the development of innovative technologies. Molecular diagnostic testing has evolved to the point where polymerase chain reaction (PCR) amplification testing for certain organisms can be performed in a routine clinical laboratory 24 hours a day, 7 days a week.1 Such methods target specific genetic regions of the organism and are used when clinical suspicion for a specific organism is present. The development of 16s rRNA gene sequencing has led to a new standard for identifying bacteria. Due to additional advances leading to ease of use, it has now become the “gold standard” for identifying bacteria, especially those traditionally thought to be difficult to identify.2 However, it is still more expensive than most traditional identification methods and is limited to use in large research and reference laboratories. One of the latest methods for bacterial identification to gain substantial momentum is matrix-assisted laser desorption ionization, coupled with time of flight mass spectrometry (MALDI-TOF MS) for identification of microorganisms.3 This technique has allowed for the rapid identification of bacteria, mycobacteria, as well as yeast.4-9 The method has proven to be cost-effective and accurate for identification of microbial organisms, obviating the need for routine biochemical methods.10 The purpose of this study was to examine the use of the MALDI-TOF MS in the clinical microbiology laboratory to identify the group of organisms commonly isolated from patients with cystic fibrosis (CF). Patients with CF are prone to develop infections with difficult to identify non-fermenting Gram negative rods including Burkholderia cepacia complex (BCC). We sought to identify these organisms in the BCC, compared to gene sequencing. To the best of our knowledge, this is the first publication looking at BCC isolates with the unabridged database. MATERIALS AND METHODS: Microbial Strains. Organisms were obtained from routine clinical specimens cultivated on a variety of different media (Remel, Lenexa, KS) including: Trypticase soy blood agar with 5% sheep blood (TSA); Chocolate agar (CHOC); MacConkey Agar (MAC); Mannitol Salt Agar (MS); and Burkholderia cepacia selective agar (BCSA). Plates were incubated in ambient air or 5% CO2 at 35-37°C and examined daily for 7 days or until all isolates were identified. Phenotypic identification of organisms conformed to routine microbiological procedures (Clinical Microbiology Procedures Handbook, ASM Press, @2010). The subset of non-fermenting gram negative rods (NFR) was identified according to manufacturer’s instructions for MicroScan (Siemens, Newark, DE) and Biomerieux API 20 NE (Durham, NC). Reference Method for Burkholderia. Definitive identification of Burkholderia spp. was obtained from genetic analyses performed at the Burkholderia cepacia Research Laboratory and Repository (University of Michigan). The methods used a combination of species-specific 16S ribosomal DNA PCR assays, recA-RFLP analysis, and recA sequence analysis as previously described.11-18 MALDI-TOF MS. MALDI-TOF MS measurements were performed on a microflex LT (Bruker Daltonic, Billerica, MA) provided with a 60-Hz nitrogen laser. Spectra were analyzed over a mass range of 2,000 to 20,000 Da. Each series of measurements was preceded by weekly calibration with a bacterial test standard (BTS 255343; Bruker Daltonic) and daily QC organisms. Routine Testing. The tops of individual colonies were transferred to 1-3 wells on a steel template carrier (96 well target). After air drying, 1 ųl of matrix solution was added to each well. The matrix was a saturated solution of α-cyano-4-hydroxycinnamic acid dissolved in a basic organic solvent composed of 50% acetonitrile and 2.5% trifluoroacetic acid. Spots were measured with the MBT FC.par flexControl method and the MBT-autoX.exe autoExecute method. All measurements resulted from six series of 40 laser shots at multiple positions on the spotted target. The bacterial spectra acquired by MALDI-TOF MS were analyzed in the MALDI BioTyper 2.0 software incorporating the MALDI BioTyper database. A score was obtained after submitting the downloaded raw spectra to the original MALDI BioTyper database version 3.0.2. This database contains spectra for 3,995 organisms, including various bacterial and fungal species. The score is a logarithmic value resulting from the alignment of the unknown raw spectrum and the best matching database spectrum. According to the specifications of the manufacturer, a high log score of >2 was required for species level identification and an intermediate log score lying between ≤2 and ≥1.7 was required for genus level identification. A low score of <1.7 was considered unreliable for identification. Extraction Procedure. An extraction procedure was performed for the single organism which demonstrated no peaks on initial testing. Briefly, approximately 3 colonies were scraped from the agar and added to 300 ųl distilled water, followed by the addition of 900 ųl of 100% ethanol. The suspension was centrifuged twice at 13,000 rpm for 2 minutes (Eppendorf Centrifuge model 5425) and the supernatant was removed. Caps were left open for 10 minutes to allow the pellet to dry. The pellet was suspended in 50 ųl of formic acid (70%) and incubated for 15 min at room temperature followed by the addition of 50 ųl of 100% acetonitrile. The mixture was centrifuged for 2 min at 13,000 rpm and 1 ųl of the supernatant was placed on 2 wells on the target. After air drying, each well was overlaid with matrix and subjected to analysis as above. Workflow Analysis. Detailed hand-motion time studies were performed on 24 respiratory specimens submitted from CF patients. Routine phenotypic tests used for identification were assessed for turn-around time and compared to MALDI-TOF MS. RESULTS: We included a total of 464 tested isolates in our analysis, representing 17 different bacterial species. Conventional identification compared to MALDI-TOF MS showed complete genus, species agreement in 92% of tested isolates, with genus agreement to 98%. The remaining 2% were either organisms with known limitations of the MALDITOF database or discrepant results. Table 1 details our findings. In summary, one hundred percent of 124 isolates of Pseudomonas aeruginosa, 189 Staphylococcus aureus, and 25 Haemophilus influenzae matched to the species level. Of 2 isolates identified by conventional methods as Ralstonia pickettii, one showed complete agreement, and the other was identified by MALDI-TOF MS as Herbaspirillum huttiense. Identification was resolved with 16S sequencing as Herbaspirillum huttiense. For 58 Stenotrophomonas maltophilia isolates, 84% showed complete genus, species agreement. The remaining 16% were identified as Pseudomonas hibiscola (n=6), Pseudomonas geniculata (n=2), and Pseudomonas betelei (n=1). MALDI-TOF MS provided accurate identification for all tested isolates of the BCC to the genus level, with 24 of 29 (83%) being speciated. These included B. cenocepacia, B. cepacia, B. multivorans and, B. vietnamiensis. In addition one isolate of B. gladioli (sensu strictu, not a member of the BCC) was accurately identified. Among the 15 tested isolates of Acinetobacter spp., 53% were accurately identified to the species level as Acinetobacter baumannii, with the remaining 47% identified to the genus level. Only one organism (a mucoid Pseudomonas aeruginosa) required the additional extraction procedure. The extraction procedure added an extra 30 minutes to final identification over the direct test method. Workflow analysis was performed to compare the hands on time, complexity, and time to result between the routine culture and biochemical methods and the MALDI-TOF MS method. A total of 47 bacterial isolates from CF respiratory cultures obtained from 24 patients were examined. When considering the additional time needed for subculturing, special stains, spot tests, quality control, and constant motion for various subculture to additional plates, MALDI-TOF MS was shown to require less technical time, was of lower complexity, and resulted in a faster time to organism identification (Graph 1). The simple, single MALDI-TOF procedure replaced 48 subcultures, 20 cetrimide plates, 25 oxidase spot tests, 19 Staphylococcus agglutination tests, 5 MicroScan identifications, and one Streptococcus agglutination test. As shown on Graph 1, among total isolates, 85% of isolates were identified by MALDI-TOF MS at 48 hours of incubation, and all were identified at five days. Using the conventional method for organism identification, only 34% were identified at 48 hours, with the remainder taking up to seven days. DISCUSSION: Using the unamended database for MALDI-TOF MS compared to routine conventional methodology for commonly identified pathogens from CF patients, we were able to show accurate results, with faster time to identification. We demonstrated complete genus, species agreement with MALDI-TOF MS compared to conventional identification in 92% of isolates, with genus agreement to 98%. One hundred percent of our tested isolates of Pseudomonas aeruginosa, Staphylococcus aureus, and Haemophilus influenzae matched to the species level. Among total isolates, 85% of organisms were identified by MALDI-TOF MS at 48 hours of incubation, compared to only 34% by conventional methods. This is consistent with other publications showing the improvement in cost and time to detection in using MALDITOF MS for routine bacterial identification.10 Others have reported the use of the MALDI-TOF MS in identification of various organisms, including yeast and mycobacteria.4-9 Degand et al used MALDI-TOF MS for identification of organisms commonly isolated from CF patients, using a modified database to include additional bacterial isolates commonly isolated from cystic fibrosis patients.19 They reported the improved identification of CF organisms when the database was supplemented with CF-specific additional reference strains. Our study is the first that we know of to identify CF organisms using the standard database with no modification. Regarding Burkholderia spp., definitive identification was achieved for B. cenocepacia, B. cepacia, B. multivorans, B. vietnamiensis and B. gladioli. This was compared to the definitive identification obtained from genetic analyses performed at the Burkholderia cepacia Research Laboratory and Repository (University of Michigan) as described in Methods. These findings are significant in that current phenotypic “kit methods” when accurate, only identify this class of organisms as Burkholderia cepacia complex. Accurate distinction between Burkholderia cepacia complex and other gram-negative non-fermenting rods is critical for appropriate management of CF patients. These organisms have been correlated with increased morbidity and mortality, may represent a contraindication to lung transplant, and require attention to infection control to prevent spread both within the hospital and the community.20, 21 Several noteworthy findings emerged from testing these organisms. Achromobacter xylosoxidans by phenotypic kit assay was commonly identified as Achromobacter ruhlandii by MALDI-TOF MS. In addition, the Acinetobacter complex of organisms identifying as Acinetobacter baumannii or A. lwoffii by MicroScan or API NE were often identified as Acinetobacter genomospecies or A. ursingii, respectively by MALDI-TOF MS. As the taxonomy of the genus Acinetobacter continues to be updated, modifications in the databases of both MALDI-TOF and phenotypic systems will be warranted. Stenotrophomonas maltophilia identified by biochemical parameters was commonly called Pseudomonas hibiscola , P. geniculata, or P. betelei by MALDI-TOF MS. These Pseudomonas species show high levels of homology with Stenotrophomonas maltophilia and are considered by some to be heterotypic synonyms.22,23 In addition, MALDI-TOF MS accurately identified Herbaspirillum huttiense, while both API NE and MicroScan identified the organism as Ralstonia pickettii. This is significant in that this organism is uncommonly isolated from CF patients, often initially identified as a member of the BCC, and is under scrutiny as a potentially significant pathogen.24 Future applications of the MALDI-TOF MS technique will continue to add to key aspects of patients care and infection control. These would include ‘standardized’ procedures for direct detection from sterile body fluids, susceptibility testing for key drugs representing major ‘drugs of choice’, and typing and fingerprinting organisms of epidemiologic significance. CONCLUSION: MALDI-TOF MS is a fast, reliable tool that can be used in a pediatric clinical microbiology laboratory. The database supplied by the company was used with no modification to assess its ability to identify organisms commonly isolated in the cystic fibrosis patient population. This is in contrast to prior studies, which used a supplemented database for identification of hard to identify bacteria. MALDI-TOF MS allows for faster identification of organisms that historically have been thought of as hard-to-identify, time-consuming, and costly. This study demonstrates that the use of MALDI-TOF MS can replace routine methods of identification for organisms cultured from CF patients. Adoption of this technique will allow for the earlier report of culture results to the clinical care team, and ultimately lead to improved patient outcomes. Part of this work was presented at the American Society for Microbiology 110th General Meeting in May of 2010 in San Diego, California. Table 1: Organisms Commonly Isolated From CF Patients: Comparison of Conventional or Reference Methods* to MALDI-TOF MS Organism No. of Isolates (% of total tested) No. (%) of Isolates ID to species ID to Genus 21(5) 11(2) 3(<1) 1(<1) 1(<1) 9(43) 8(73) 12(57) 3(27) 3(100) 1(100) 1(100) Ralstonia pickettii 2(<1) 1(50) 1(50)1 Stenotrophomonas maltophilia 58(13) 49(84) 9(16)2 Pseudomonas aeruginosa 3 124(27) 124(100) B.cepacia complex Burkholderia cenocepacia* Burkholderia cepacia* Burkholderia contaminans* Burkholderia dolosa* Burkholderia multivorans* Burkholderia vietnamiensis* 14(3) 2(<1) 1(<1) 1(<1) 8(<2) 2(<1) 12(86) 1(50) Burkholderia gladioli* 1(<1) 1(100) 25(5) 189(41) 25(100) 189(100) Non-fermenters Achromobacter xylosoxidans Acinetobacter baumanii Acinetobacter lwoffii Acinetobacter sp. Pandoraea pnomenusa* Other Routine Organisms Haemophilus influenzae Staphylococcus aureus 8(100) 2(100) * Reference methods performed at the Burkholderia cepacia Research Laboratory and Repository 1 Strain identified by MALDI-TOF MS as Herbaspirillum huttiense confirmed by 16s sequence analysis 2 Known limitation of taxon overlap with Pseudomonas hibiscola (n=6), P. geniculata (n=2) and P. betelei 3 One mucoid isolate required extraction 2(14) 1(50) 1(100) 1(100) No Match Graph 1. Time to Identification, MALDI-TOF MS vs. Conventional Methods 25 Total Isolates 20 CONVENTIONAL MALDI-TOF 15 10 5 0 1 2 3 4 Time (Days) 5 6 7 Graph 1. Comparison of MALDI-TOF MS to conventional methods for time to identification. 47 total isolates identified from 24 cystic fibrosis patients. Pathogens identified include Pseudomonas aeruginosa, Staphylococcus aureus, Stenotrophomonas maltophilia, Achromobacter xylosoxidans, Burkholderia cepacia, Group A streptococcus, and Klebsiella oxytoca. References: 1 Kost CB et al. Multicenter Beta Trial of the GeneXpert Enterovirus Assay. J. Clin. Microbiol., 2007; 45: 1081–1086. 2 Clarridge JE. Impact of 16S rRNA Gene Sequence Analysis for Identification of Bacteria on Clinical Microbiology and Infectious Diseases. Clin. Micro. Rev., 2004; 17(4): 840-862. 3 Seng P et al. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin. Infect. Dis., 2009; 49 (4): 543–551. 4 Marklein G et al. Matrix-assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry for Fast and Reliable Identification of Clinical Yeast Isolates. J. Clin. Microbiol., 2009; 47: 2912-2917. 5 Dhiman N et al. Performance and Cost Analysis of Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry for Routine Identification of Yeast. J. Clin. Microbiol., 2011; 49: 1614-1616. 6 Pignone M et al. Identification of Mycobacteria by Matrix-Assisted Laser Desorption Ionization -- Timeof-Flight Mass Spectrometry. J. Clin. Microbiol., 2006; 44: 1963-1970. 7 Lotz A et al. Rapid Identification of Mycobacterial Whole Cells in Solid and Liquid Culture Media by Matrix-assisted Laser Desorption Ionization – Time of Flight Mass Spectrometry. J. Clin. Microbiol., 2010; 48: 4481-4486. 8 Saleeb PG et al. Identification of Mycobacteria in Solid-Culture Media by Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry. J. Clin. Microbiol., 2011; 49: 1790-1794. 9 Bizzini A et al. Performance of Matrix -Assisted Laser Desoption Ionization –Time of Flight Mass Spectroscopy for Identification of Bacterial Strains Routinely Isolated in a Clinical Microbiology Laboratory. J. Clin. Microbiol., 2010; 48: 1549-1554. 10 Gaillot O et al. Cost-effectiveness of Switching to MALDI-TOF MS for Routine Bacterial Identification. J. Clin. Microbiol. 2011. 11 LiPuma JJ et al. Development of rRNA-based PCR assays for identification of Burkholderia cepacia complex isolates recovered from cystic fibrosis patients. J. Clin. Microbiol., 1999; 37(10): 3167-3170. 12 Vermis K et al. Evaluation of species specific recA-based PCR tests for genomovar level identification within the Burkholderia cepacia complex. J. Med. Microbiol., 2002; 51(11): 937-940. 13 Whitby PW et al. Species specific PCR as a tool for the identification of Burkholderia gladioli. J. Clin. Microbiol., 2000; 38: 282-285. 14 Whitby PW et al. Identification and detection of Stenotrophomonas maltophilia by rRNA-directed PCR. J. Clin. Microbiol., 2000; 38: 4305-4309. 15 Coenye T et al. Identification of Pandoraea species by 16S ribosomal DNA-based PCR assays. J. Clin. Microbiol., 2001; 39:4452-4455. 16 Liu L et al. Ribosomal DNA-directed PCR for identification of Achromobacter (Alcaligenes) xylosoxidans recovered from sputum samples from cystic fibrosis patients. J. Clin. Microbiol., 2002; 40:1210-1213. 17 Coenye T et al. Infection by Ralstonia species in cystic fibrosis patients: Identification of R. pickettii and R. mannitolilytica by polymerase chain reaction. Emerg. Infect. Dis., 2002; 8:692-696. 18 Coenye T et al. Use of PCR analyses to define the distribution of Ralstonia species recovered from patients with cystic fibrosis. J. Clin. Microbiol., 2005; 43:3463-3466. 19 Degand N et al. MALDI-TOF MS for Identification of Nonfermenting Gram-negative Bacilli Isolated from Cystic Fibrosis Patients. J. Clin. Microbiol., 2008; 46: 3361-3367. 20 Corerey M and Farewell V. Determinants of mortality from cystic fibrosis in Cannada. 1070-1989. Am. J. Epidemiol., 1996; 143:1007-1017. 21 Lipuma JJ. Update on the Burkholderia cepacia complex. Curr. Opin. Pulm. Med. 2005; 11:528-533. 22 Van Den Mooter M, Swings J. Numerical analysis of 295 phenotypic features of 266 Xanthomonas strain and related strains and improved taxonomy of the genus. Int. J. Syst. Bacteriol. 1990; 40:348-369. 23 Anzai Y et al. Phylogenetic affiliation of the pseudomonads based on 16S rRNA sequence. Int. J. System. Evol. Microbiol. 2000; 50:1563-1589. 24 Spilker T et al. Recovery of Herbaspirillum Species from Persons with Cystic Fibrosis. J. Clin. Microbiol., 2008; 46: 2774-2777.