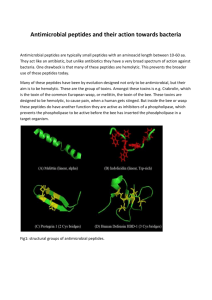
bit25003-sm-0001-SuppData
advertisement

Supplementary Material Materials and Methods CD spectroscopy All far-UV circular dichroism (CD) measurements were recorded in a ChirascanTM Circular Dichrosim spectrometer (Applied Photophysics Ltd., UK) using a 0.01 cm path length cuvette (Hellma). Spectra data were collected with a step size of 0.5 and time constant of 0.5 s. The peptide concentration for the measurements was fixed at 100 g/ml in 10 mM phosphate buffer, pH 7.2. The peptide’s secondary structural change upon interaction with lipids was recorded in the same buffer containing either 10 mM SDS or DPC. All spectra were recorded at 25°C from 190 to 240 nm using a bandwidth of 1 nm and averaged over three scans. Baseline scans were obtained using the same parameters for buffer and micelles, and subtracted from the respective data scans with peptides. The final corrected averaged spectra were expressed in molar ellipticity. Results and Discussions CD analysis of the peptides showed that in the free state, all the peptides demonstrated a broad negative band at ~200 nm, indicative of unordered state, and a second smaller negative band at 227 nm (Supplementary Figure 1A). The band at 227 nm has been reported to arise due to stacking of the tryptophan aromatic rings and interaction with the peptide backbone (Woody 1994, Friedrich, Rozek et al. 2001). The absence of the 227 nm band in D1 suggests the occurrence of aromatic ring interactions in other W-containing peptides D2 to D6. When bound to the negatively charged SDS micelles, all amidated peptides except D5 retained both the negative bands at 200 and 227 nm. Amidated D5 which contains three W residues appears to show a mixed helical structure in SDS and DPC, which could also be contributed by the 1 tryptophan interference at wavelength 220-240 nm (Supplementary Figure 1B and 1C). Taken together, the secondary structure analysis of the peptides in the detergent micelles suggests that none of the peptides demonstrate spectra indicative of defined secondary structural elements. The addition of salt up to 150mM NaCl, did not alter the CD spectrum of the active peptides, D5 and D6, in DPC micelles, indicating that salt does not interfere with peptide backbone conformation (Supplementary Figure 1D). Supplementary Figure 1 Figure S1 Secondary structure analysis of the amidated decamers in solution and upon interaction with lipid micelles. Far UV circular dichroism spectra of the peptides in 10 mM phosphate buffer, pH 7.2 (panel A), negatively charged SDS (panel B), zwitterionic DPC 2 micelles (panel C) and DPC bound D5-NH2 and D6-NH2 in the presence and absence of salt (panel D) recorded at 25° C. Supplementary Figure 2 Figure S2 Overlay of the selected sections of the 2-D NOESY spectra of D5-NH2 (panel A) and D6-NH2 (panel B) in 10 mM sodium phosphate buffer (cyan contour) and in solution containing 200 mM SDS (red contour) showing NOE correlations among down field shifted amide and aromatic resonances (along 2) with upfield shifted resonances (along 1). 3 Supplementary Table 1 Summary of the structural statistics of twenty lowest energy structures of D5 and D6 in SDS micelles calculated using CYANA. Distance restraints D5 D6 intraresidue (|i-j| =0) 36 27 sequential (|i-j| =1) 26 26 medium range (2≤|i-j|≤4) 10 9 total NOE constraints 72 62 backbone atoms (N, Cα, C`) (Å) 0.66 0.65 all heavy atoms (Å) 1.97 2.02 Deviation from the mean structure 4