tpj12011-sup-0019-SupportingInformationlegends
advertisement

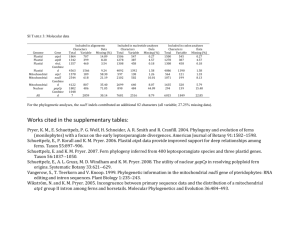
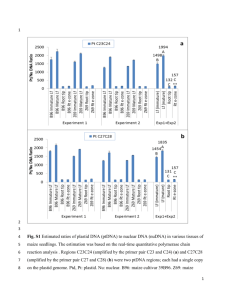
1 Supporting Information legends Figure S1. Plastid transcription is required for PhANG expression in Arabidopsis. qPCR analysis of steady state mRNA transcript levels of 2-day old etiolated seedlings grown in the presence or absence of 50 g/ml rifampicin. Data shown are the mean +/- SEM of triplicate reactions of a representative experiment. Figure S2. SIG2 and SIG6-mediated plastid transcription is required for PhANG expression. qPCR analysis of steady state a) SIG, b) NEP-dependent plastid, c) control genes, d) PEP-dependent, e) and PhANG mRNA transcript levels of 2-day old seedlings. Data shown are the mean +/- SEM of triplicate reactions of a representative experiment. f) Chlorophyll levels of three-day-old seedlings. Data shown are the +/- SEM of biological duplicates. g) Image of 3 day-old complemented sig mutant seedlings. h) SIG2- and SIG6-YFP-HA localize to the chloroplasts. Florescence was imaged by confocal microscopy and representative images are shown. Scale bars = 20 M. Figure S3. Genetic interactions between sigma and gun mutants. qPCR analysis of steady state plastid mRNA transcript levels of two-day-old a) etiolated and b) de-etiolated seedlings. Data shown are the mean +/- SEM of triplicate reactions of a representative experiment. c) Images of seven day-old seedlings grown on medium supplemented with 1% sucrose. Figure S4. Correlation of PhANG and plastid gene expression in sig2 and sig6. a) qPCR time course experiment of PhANG (fist and second row), plastid (third and fourth row), and SIG (fifth and sixth row) transcripts in the wt, sig2, and sig6 mutants. Data shown are the mean +/- SEM of triplicate reactions of a representative experiment. b) Western blot analysis of plastid proteins. 20 g of total protein was fractioned by SDS-PAGE and blotted. Filters were probed with antibodies specific for the proteins indicated to the left. Figure S5. SIG2 and SIG6 are partially redundant for genome coordination. a) qPCR time course experiment of nuclear SIG transcripts in the wt, sig2, sig6, and sig2/sig6 2 mutants. Shown are the mean +/- SEM of triplicate reactions of a representative experiment. b) Image of five-week-old sig mutant seedlings grown on medium supplemented with 1% sucrose. Figure S6. The gun1 mutation further reduces plastid transcripts in sig6 mutants. Analysis of steady-state transcript levels of individual plastid genes in two-day-old seedlings as determined by strand-specific RNA-sequence analysis. Shown are the relative expression levels of all monitored plastid genes between a) wt vs. sig6 and b) sig6 vs. sig6/gun1. c) Correlation of independent strand-specific RNA-sequence and RTqPCR analyses. The fold change values (log2) (sig6 vs. wt) of ten plastid genes (rbcL, rpoC1, ndhC, clpP, psbB, ndhF, psaJ, psbA, psbD, and psbN) are plotted and shown to exhibit a high correlation (r2=0.901). Figure S7. Analysis of glutamyl-tRNA levels in sig2 and sig6 mutants. Northern blot analysis of glutamyl-tRNA from two-day-old seedlings. Figure S8. SIG2-mediated signals involve tetrapyrrole synthesis. qPCR analysis of steady state a) PEP-dependent plastid and b) FC mRNA transcript levels of two-day-old seedlings. Data shown are the mean +/- SEM of triplicate reactions of a representative experiment. c) Western blot analysis of overproduced tagged ferrochelatase proteins. 20 µg of total protein was fractioned by SDS-PAGE and blotted. Filters were probed with an HA antibody. d) Images of ALA-fed seedlings after two days of 24h white light. Figure S9. NF reduces plastid gene expression. qPCR analysis of steady state plastid mRNA transcript levels of two-day-old seedlings. Data shown are the mean +/- SEM of triplicate reactions of a representative experiment. Figure S10. Microarray analysis of sigma mutants and herbicide inhibitor treatments. Correlation of independent microarray and RT-qPCR analyses. The fold change values (log2) of eight plastid genes (LHCB1.2, CA1, PC, CBL, PIR, PPI2, SIG2, AND SIG6) are plotted and shown for a) wt vs. wt + NF, b) wt vs. wt + Linc, c) wt vs. sig2, d) and wt vs. 3 sig6. Shown are 4-way Venn diagrams depicting the overlap of genes e) repressed or f) induced by sig2, sig6, NF treatment, and Linc treatment. Table S1. Arabidopsis mutants used in study. Table S2. List of retrograde-insensitive control genes used in this study. Table S3. Primers used in study. Table S4. Plasmids used in study. Table S5. Overlap of retrograde-responsive nuclear genes identified by microarray analysis. Table S6. List of genes up-regulated by retrograde signals as identified by microarray analysis. Table S7. List of genes down-regulated by retrograde signals as identified by microarray analysis. Methods S1. Supplemental experimental procedures: construction of overexpression plasmids; protein extraction and Western Blot analysis; microarray expression and cluster analysis.