Student Worksheet
advertisement
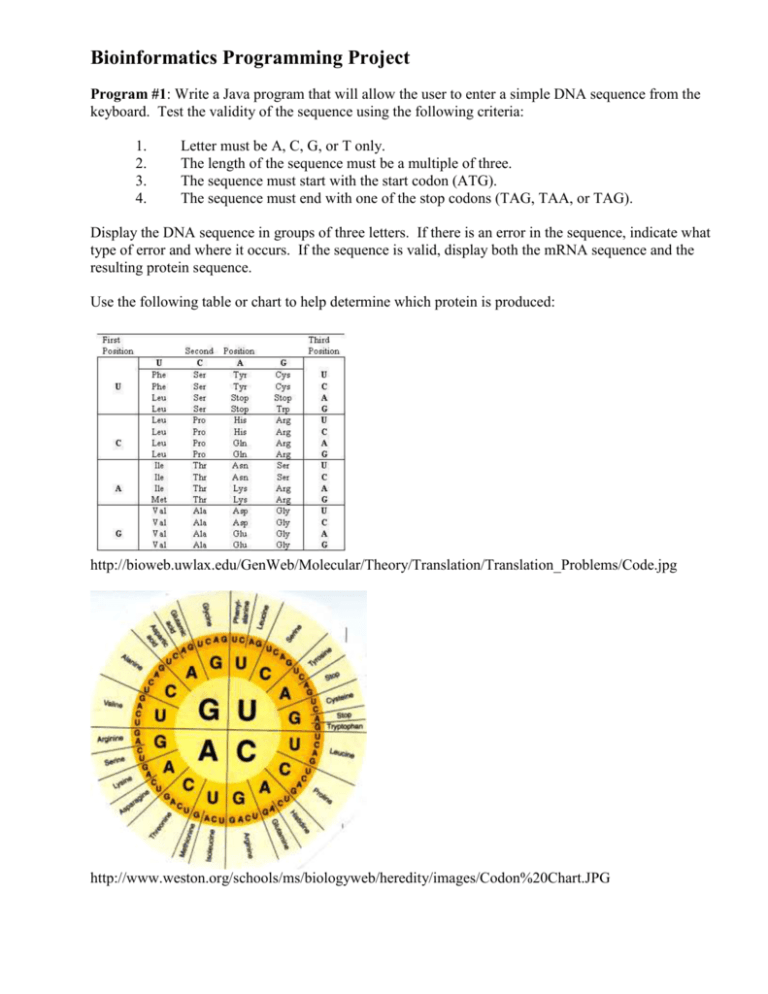
Bioinformatics Programming Project Program #1: Write a Java program that will allow the user to enter a simple DNA sequence from the keyboard. Test the validity of the sequence using the following criteria: 1. 2. 3. 4. Letter must be A, C, G, or T only. The length of the sequence must be a multiple of three. The sequence must start with the start codon (ATG). The sequence must end with one of the stop codons (TAG, TAA, or TAG). Display the DNA sequence in groups of three letters. If there is an error in the sequence, indicate what type of error and where it occurs. If the sequence is valid, display both the mRNA sequence and the resulting protein sequence. Use the following table or chart to help determine which protein is produced: http://bioweb.uwlax.edu/GenWeb/Molecular/Theory/Translation/Translation_Problems/Code.jpg http://www.weston.org/schools/ms/biologyweb/heredity/images/Codon%20Chart.JPG Program #2: Write a Java program that will allow the user to a select text file that contains a DNA sequence. Use the same criteria from Program #1 to text for validity. Because of the potential long length of the sequence, display the DNA sequence in groups of 99 letters per line and the protein sequences in groups of 33 letters per line. Program #3: Write a Java program that will allow the user to enter a DNA sequence from the keyboard or select a text file that contains a DNA sequence. Once the DNA sequence has been read into your program, cause a random mutation at a random location. (Remembering that biologist start counting at 1 instead of 0). Compare the resulting protein sequence with that of the original sequence. Determine which of the following mutations occurred: silent (no change in the protein), missense (a change in the protein), nonsense (stops the code), or frame shift (all letter in the DNA sequence are shifted to the left one location). Also display the location of the mutation and what was changed in the sequence. Program #4: Write a Java program to allow the user to enter a DNA sequence from the keyboard. Then allow the user to select a text file that contains a DAN sequence. Determine whether or not the sequence that was entered exist file’s sequence. If it exists, indicate the starting location of each occurrence of the given sequence.