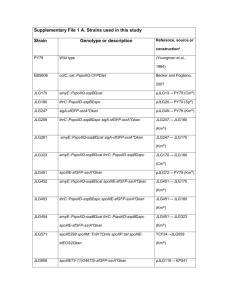
Quantitative determinants of aerobic glycolysis identify flux through
advertisement

Supplementary material and methods Kinetic mass-balance equations Intracellular metabolites: 𝑑𝐺𝐿𝐶𝑖 = 𝐽𝑔𝑡𝑟 − 𝑉ℎ𝑘 𝑑𝑡 𝑑𝐺6𝑃 𝑑𝑡 𝑑𝐹6𝑃 𝑑𝑡 𝑑𝐹𝐵𝑃 𝑑𝑡 = 𝑉ℎ𝑘 − 𝑉𝑔𝑝𝑖 − 𝑉𝑔𝑠𝑦𝑛 − 𝑉𝑔6𝑝𝑑 + 𝑉𝑔𝑝ℎ𝑜𝑠 = 𝑉𝑔𝑝𝑖 − 𝑉𝑝𝑓𝑘 = 𝑉𝑝𝑓𝑘 − 𝑉𝑎𝑙𝑑 𝑑𝐺𝐴3𝑃 = 𝑉𝑎𝑙𝑑 − 𝑉𝑔𝑎𝑝𝑑ℎ + 𝑉𝑡𝑝𝑖 𝑑𝑡 𝑑𝐷𝐻𝐴𝑃 𝑑𝑡 𝑑13𝐵𝑃𝐺 𝑑𝑡 𝑑3𝑃𝐺 𝑑𝑡 𝑑2𝑃𝐺 𝑑𝑡 𝑑𝑃𝐸𝑃 𝑑𝑡 𝑑𝑃𝑌𝑅 𝑑𝑡 𝑑𝐿𝐴𝐶𝑖 𝑑𝑡 = 𝑉𝑎𝑙𝑑 − 𝑉𝑡𝑝𝑖 − 𝑉𝑔𝑟𝑝𝑑ℎ − 𝑉𝑔𝑝𝑠 = 𝑉𝑔𝑎𝑝𝑑ℎ − 𝑉𝑝𝑔𝑘 = 𝑉𝑝𝑔𝑘 − 𝑉𝑝𝑔𝑚 − 𝑉𝑝𝑔𝑑ℎ = 𝑉𝑝𝑔𝑚 − 𝑉𝑒𝑛 = 𝑉𝑒𝑛 − 𝑉𝑝𝑘 = 𝑉𝑝𝑘 − 𝑉𝑜𝑥𝑝ℎ𝑜𝑠 − 𝑉𝑙𝑑ℎ − 𝑉𝑎𝑙𝑡 = 𝑉𝑙𝑑ℎ − 𝐽𝑙𝑡𝑟 𝑑𝐴𝑇𝑃 = −𝑉ℎ𝑘 −𝑉𝑝𝑓𝑘 +𝑉𝑝𝑔𝑘 + 𝑉𝑝𝑘 + 𝑛𝑜𝑝 𝑉𝑜𝑥𝑝ℎ𝑜𝑠 − 𝑉𝑎𝑡𝑝𝑎𝑠𝑒 + 𝑉𝑐𝑘 − 𝑉𝑎𝑘 + 𝑛𝑓𝑎𝑑 𝑉𝑔𝑝𝑠 + 𝑛𝑛𝑎𝑑 𝑉𝑚𝑎𝑠 𝑑𝐴𝐷𝑃 = 𝑉ℎ𝑘 +𝑉𝑝𝑓𝑘 −𝑉𝑝𝑔𝑘 − 𝑉𝑝𝑘 − 𝑛𝑜𝑝 𝑉𝑜𝑥𝑝ℎ𝑜𝑠 + 𝑉𝑎𝑡𝑝𝑎𝑠𝑒 − 𝑉𝑐𝑘 + 2 ⋅ 𝑉𝑎𝑘 − 𝑛𝑓𝑎𝑑 𝑉𝑔𝑝𝑠 − 𝑑𝑡 𝑛𝑛𝑎𝑑 𝑉𝑚𝑎𝑠 𝑑𝑡 𝑑𝑃𝑖 𝑑𝑡 = −𝑉𝑔𝑎𝑝𝑑ℎ + 𝑉𝑝𝑔𝑑ℎ − 𝑛𝑜𝑝 𝑉𝑜𝑥𝑝ℎ𝑜𝑠 + 𝑉𝑎𝑡𝑝𝑎𝑠𝑒 − 𝑛𝑛𝑎𝑑 𝑉𝑚𝑎𝑠 + 2 ⋅ 𝑉𝑔𝑠𝑦𝑛 −𝑉𝑔𝑝ℎ𝑜𝑠 𝑑𝑁𝐴𝐷𝐻 𝑑𝑡 = 𝑉𝑔𝑎𝑝𝑑ℎ − 𝑉𝑙𝑑ℎ + 𝑉𝑝𝑔𝑑ℎ − 𝑉𝑔𝑟𝑝𝑑ℎ − 𝑉𝑚𝑎𝑠 −𝑉𝑔𝑝𝑠 𝑑𝑃𝐶𝑟 𝑑𝑡 𝑑𝑂2𝑖 𝑑𝑡 = −𝑉𝑐𝑘 = 𝐽𝑂2 − 𝑛𝑜𝑥 𝑉𝑜𝑥𝑝ℎ𝑜𝑠 𝑑𝐺𝐿𝐺 = 𝑉𝑔𝑠𝑦𝑛 − 𝑉𝑔𝑝ℎ𝑜𝑠 𝑑𝑡 𝑑𝑆𝐸𝑅 𝑑𝑡 𝑑𝐺𝐿𝑌 𝑑𝑡 = 𝑉𝑝𝑔𝑑ℎ − 𝑉𝑔ℎ𝑚𝑡 − 𝑉𝑠𝑝𝑡 = 𝑉𝑔ℎ𝑚𝑡 − 𝑉𝑔𝑝𝑡 ; Extracellular metabolites: 𝑑𝐺𝐿𝐶𝑒 𝑑𝑡 𝑑𝐿𝐴𝐶𝑒 𝑑𝑡 = 0 = 𝑟𝑖𝑒 ⋅ 𝐽𝑙𝑡𝑟 Rate laws for each flux 1. Glucose transport 𝐽𝑔𝑡𝑟 = 𝐺𝑙𝑐𝑖 𝐺𝑙𝑐𝑒 − ) 𝐾𝑚𝑔𝑙𝑐 𝐾𝑚𝑔𝑙𝑐 𝐺𝑙𝑐𝑖 𝐺𝑙𝑐𝑒 1+ + 𝐾𝑚𝑔𝑙𝑐 𝐾𝑚𝑔𝑙𝑐 𝑉𝑚𝑎𝑥 𝑡𝑟 ( 2. Glucose utilization 𝐺𝐿𝐶𝑖 ∗𝐴𝑇𝑃 𝑚𝑎𝑥 𝐺6𝑃∗𝐴𝐷𝑃 −𝑉𝑟ℎ𝑘 𝐾𝑚ℎ𝑘𝑓 𝐾𝑚ℎ𝑘𝑟 𝐺𝐿𝐶𝑖 ∗𝐴𝑇𝑃 𝐺6𝑃∗𝐴𝐷𝑃 1+ + 𝐾𝑚ℎ𝑘𝑓 𝐾𝑚ℎ𝑘𝑟 𝑚𝑎𝑥 𝑉𝑓ℎ𝑘 𝑉ℎ𝑘 = ( )∗ 𝑖 𝐾ℎ𝑘 ⁄ 𝑖 (𝐾ℎ𝑘 + 𝐺6𝑃) 3. Phosphoglucose isomerase activity a. 𝑉𝑝𝑔𝑖 = ( 𝐺6𝑃 𝑚𝑎𝑥 𝐹6𝑃 −𝑉𝑟𝑝𝑔𝑖 𝐾𝑚𝑝𝑔𝑖𝑓 𝐾𝑚𝑝𝑔𝑖𝑟 𝐺6𝑃 𝐹6𝑃 1+ + 𝐾𝑚𝑝𝑔𝑖𝑓 𝐾𝑚𝑝𝑔𝑖𝑟 𝑚𝑎𝑥 𝑉𝑓𝑝𝑔𝑖 ) 4. Phosphofructokinase 𝑉𝑝𝑓𝑘 = ( + 𝐹6𝑃∗𝐴𝑇𝑃 𝑚𝑎𝑥 𝐹16𝐵𝑃∗𝐴𝐷𝑃∗𝐻 −𝑉𝑟𝑝𝑓𝑘 𝐾𝑚𝑝𝑓𝑘𝑓 𝐾𝑚𝑝𝑓𝑘𝑟 𝐹6𝑃∗𝐴𝑇𝑃 𝐹16𝐵𝑃∗𝐴𝐷𝑃∗𝐻+ 1+ + 𝐾𝑚𝑝𝑓𝑘𝑓 𝐾𝑚𝑝𝑓𝑘𝑟 𝑚𝑎𝑥 𝑉𝑓𝑝𝑓𝑘 5. Aldolase 𝑉𝑎𝑙 = 𝐹16𝐵𝑃 𝑚𝑎𝑥 𝐺𝐴𝑃∗𝐷𝐻𝐴𝑃 −𝑉𝑟𝑎𝑙 𝐾𝑚𝑎𝑙𝑓 𝐾𝑚𝑎𝑙𝑟 𝐹16𝐵𝑃 𝐺𝐴𝑃∗𝐷𝐻𝐴𝑃 1+ + 𝐾𝑚𝑎𝑙𝑓 𝐾𝑚𝑎𝑙𝑟 𝑚𝑎𝑥 𝑉𝑓𝑎𝑙 6. Triosephosphate isomerase ) ∗ 𝐴𝑀𝑃⁄ 𝑎 (𝐾𝑝𝑓𝑘 + 𝐴𝑀𝑃) 𝐷𝐻𝐴𝑃 𝑚𝑎𝑥 𝐺𝐴𝑃 −𝑉𝑟𝑡𝑝𝑖 𝐾𝑚𝑡𝑝𝑖𝑓 𝐾𝑚𝑡𝑝𝑖𝑟 𝐷𝐻𝐴𝑃 𝐺𝐴𝑃 1+ + 𝐾𝑚𝑡𝑝𝑖𝑓 𝐾𝑚𝑡𝑝𝑖𝑟 𝑚𝑎𝑥 𝑉𝑓𝑡𝑝𝑖 𝑉𝑡𝑝𝑖 = 7. Glyceraldehyde phosphate dehydrogenase 𝑉𝑔𝑎𝑝𝑑ℎ = + 𝐺𝐴𝑃∗𝑃𝑖 ∗𝑁𝐴𝐷+ 𝑚𝑎𝑥 13𝐵𝑃𝐺∗𝑁𝐴𝐷𝐻∗𝐻 −𝑉𝑟𝑔𝑎𝑝𝑑ℎ 𝐾𝑚𝑔𝑎𝑝𝑑ℎ𝑓 𝐾𝑚𝑔𝑎𝑝𝑑ℎ𝑟 𝑃 𝐺𝐴𝑃 𝑁𝐴𝐷+ 𝐺𝐴𝑃∗𝑃𝑖 ∗𝑁𝐴𝐷+ 13𝐵𝑃𝐺 𝑁𝐴𝐷𝐻 13𝐵𝑃𝐺∗𝑁𝐴𝐷𝐻∗𝐻+ 1+ 𝑔𝑎𝑝𝑑ℎ + 𝑖 + 𝑔𝑎𝑝𝑑ℎ + + + 𝑔𝑎𝑝𝑑ℎ + 𝐾𝑚𝑝𝑖 𝐾𝑚𝑔𝑎𝑝𝑑ℎ𝑓 𝐾𝑚𝑏𝑝𝑔 𝐾𝑚𝑔𝑎𝑝𝑑ℎ𝑟 𝐾𝑚𝑔𝑎𝑝 𝐾 𝐾 𝑚𝑛𝑎𝑑 𝑚𝑛𝑎𝑑ℎ 𝑚𝑎𝑥 𝑉𝑓𝑔𝑎𝑝𝑑ℎ , 8. Phosphoglycerate kinase 𝑚𝑎𝑥 13𝐵𝑃𝐺 ∗ 𝐴𝐷𝑃 𝑚𝑎𝑥 3𝑃𝐺 ∗ 𝐴𝑇𝑃 𝑉𝑓𝑝𝑔𝑘 − 𝑉𝑟𝑝𝑔𝑘 𝐾𝑚𝑝𝑔𝑘𝑓 𝐾𝑚𝑝𝑔𝑘𝑟 𝑉𝑝𝑔𝑘 = 13𝐵𝑃𝐺 ∗ 𝐴𝐷𝑃 3𝑃𝐺 ∗ 𝐴𝑇𝑃 1+ + 𝐾𝑚𝑝𝑔𝑘𝑓 𝐾𝑚𝑝𝑔𝑘𝑟 9. Phosphoglycerate mutase 𝑉𝑝𝑔𝑚 = 3𝑃𝐺 2𝑃𝐺 𝑚𝑎𝑥 −𝑉𝑟𝑝𝑔𝑚 𝐾𝑚𝑝𝑔𝑚𝑓 𝐾𝑚𝑝𝑔𝑚𝑟 3𝑃𝐺 2𝑃𝐺 1+ + 𝐾𝑚𝑝𝑔𝑚𝑓 𝐾𝑚𝑝𝑔𝑚𝑟 𝑚𝑎𝑥 𝑉𝑓𝑝𝑔𝑚 10. Enolase 𝑉𝑒𝑛 = 2𝑃𝐺 𝑚𝑎𝑥 𝑃𝐸𝑃 −𝑉𝑟𝑒𝑛 𝐾𝑚𝑒𝑛𝑓 𝐾𝑚𝑒𝑛𝑟 2𝑃𝐺 𝑃𝐸𝑃 1+ + 𝐾𝑚𝑒𝑛𝑓 𝐾𝑚𝑒𝑛𝑟 𝑚𝑎𝑥 𝑉𝑓𝑒𝑛 11. Pyruvate kinase 𝑚𝑎𝑥 𝑃𝐸𝑃 ∗ 𝐴𝐷𝑃 𝑚𝑎𝑥 𝑃𝑌𝑅 ∗ 𝐴𝑇𝑃 𝑉𝑓𝑝𝑘 − 𝑉𝑟𝑝𝑘 𝐾𝑚𝑝𝑘𝑓 𝐾𝑚𝑝𝑘𝑟 𝑉𝑝𝑘 = ( ) ∗ 𝐹16𝐵𝑃⁄ 𝑎 𝑃𝐸𝑃 ∗ 𝐴𝐷𝑃 𝑃𝑌𝑅 ∗ 𝐴𝑇𝑃 (𝐾𝑝𝑘 + 𝐹16𝐵𝑃) 1+ + 𝐾𝑚𝑝𝑘𝑓 𝐾𝑚𝑝𝑘𝑟 12. Lactate dehydrogenase 𝑉𝑙𝑑ℎ = + 𝑃𝑌𝑅∗𝑁𝐴𝐷𝐻 𝑚𝑎𝑥 𝐿𝐴𝐶𝑖 ∗𝑁𝐴𝐷 −𝑉𝑟𝑙𝑑ℎ 𝐾𝑚𝑙𝑑ℎ𝑓 𝐾𝑚𝑙𝑑ℎ𝑟 𝑃𝑌𝑅 𝑁𝐴𝐷𝐻 𝑃𝑌𝑅∗𝑁𝐴𝐷𝐻 𝐿𝐴𝐶𝑖 𝑁𝐴𝐷+ 𝐿𝐴𝐶𝑖 ∗𝑁𝐴𝐷+ 1+ 𝑙𝑑ℎ + 𝑙𝑑ℎ + + 𝑙𝑑ℎ + 𝑙𝑑ℎ + 𝐾𝑚𝑙𝑑ℎ𝑓 𝐾𝑚𝑙𝑑ℎ𝑟 𝐾𝑚𝑝𝑦𝑟 𝐾𝑚𝑛𝑎𝑑ℎ 𝐾𝑚𝑙𝑎𝑐 𝐾𝑚𝑛𝑎𝑑 𝑚𝑎𝑥 𝑉𝑓𝑙𝑑ℎ 13. Lactate Transport 𝐽𝑙𝑡𝑟 = 𝐿𝐴𝐶𝑖 𝐿𝐴𝐶𝑒 − ) 𝐾𝑚𝑙𝑎𝑐 𝐾𝑚𝑙𝑎𝑐 𝐿𝐴𝐶𝑖 𝐿𝐴𝐶𝑒 1+ + 𝐾𝑚𝑙𝑎𝑐 𝐾𝑚𝑙𝑎𝑐 𝑚𝑎𝑥 𝑉𝑙𝑡𝑟 ( 14. OxPhos 𝑉𝑜𝑥 = 𝑃𝑌𝑅 𝐾𝑚𝑝𝑦𝑟𝑜𝑥 𝑃𝑌𝑅 1+ 𝐾𝑚𝑝𝑦𝑟𝑜𝑥 𝑚𝑎𝑥 𝑉𝑜𝑥 * 𝑂2𝑖 𝐾𝑚𝑜2𝑖 𝑂 1+ 2𝑖 𝐾𝑚𝑜2𝑖 ∗ 𝐴𝐷𝑃 𝐾𝑚𝑎𝑑𝑝𝑜𝑥 𝐴𝐷𝑃 1+ 𝐾𝑚𝑎𝑑𝑝𝑜𝑥 ; 15. Malate-Aspartate shuttle 𝑉𝑚𝑎𝑠 = 𝑉𝑜𝑥 = 𝑃𝑌𝑅 𝐾𝑚𝑝𝑦𝑟𝑜𝑥 𝑃𝑌𝑅 1+ 𝐾𝑚𝑝𝑦𝑟𝑜𝑥 𝑚𝑎𝑥 𝑉𝑜𝑥 ∗ 𝑂2𝑖 𝐾𝑚𝑜2𝑖 𝑂 1+ 2𝑖 𝐾𝑚𝑜2𝑖 ∗ 𝐴𝐷𝑃 𝐾𝑚𝑎𝑑𝑝𝑜𝑥 𝐴𝐷𝑃 1+ 𝐾𝑚𝑎𝑑𝑝𝑜𝑥 16. ATPase + 𝐴𝑇𝑃 𝑚𝑎𝑥 𝐴𝐷𝑃∗𝑃𝑖 ∗𝐻 −𝑉𝑟𝑎𝑡𝑝 𝐾𝑚𝑎𝑡𝑝𝑓 𝐾𝑚𝑎𝑡𝑝𝑟 𝐴𝐷𝑃∗𝑃𝑖 ∗𝐻+ 𝐴𝑇𝑃 1+ + 𝐾𝑚𝑎𝑡𝑝𝑓 𝐾𝑚𝑎𝑡𝑝𝑟 𝑚𝑎𝑥 𝑉𝑓𝑎𝑡𝑝 𝑉𝑎𝑡𝑝𝑎𝑠𝑒 = 17. Creatine kinase 𝑉𝑐𝑘 = 𝑃𝐶𝑅∗𝐴𝐷𝑃 𝑚𝑎𝑥 𝐶𝑅∗𝐴𝑇𝑃 −𝑉𝑟𝑐𝑘 𝐾𝑚𝑐𝑘𝑓 𝐾𝑚𝑐𝑘𝑟 𝑃𝐶𝑅∗𝐴𝐷𝑃 𝐶𝑅∗𝐴𝑇𝑃 1+ + 𝐾𝑚𝑐𝑘𝑓 𝐾𝑚𝑐𝑘𝑟 𝑚𝑎𝑥 𝑉𝑓𝑐𝑘 18. Adenylate kinase 𝑉𝑎𝑘 = 2 𝐴𝑇𝑃∗𝐴𝑀𝑃 𝑚𝑎𝑥 𝐴𝐷𝑃 −𝑉𝑟𝑎𝑘 𝐾𝑚𝑎𝑘𝑓 𝐾𝑚𝑎𝑘𝑟 𝐴𝑇𝑃∗𝐴𝑀𝑃 𝐴𝐷𝑃2 1+ + 𝐾𝑚𝑎𝑘𝑓 𝐾𝑚𝑎𝑘𝑟 𝑚𝑎𝑥 𝑉𝑓𝑎𝑘 19. Oxygen transport to the cell 𝐽𝑂𝑡𝑟2 = 𝑘𝑂2 (𝐶𝑂2𝑒 − 𝐶𝑂2𝑖 ) 20. De novo synthesis of Serine through 3-phosphoglycerate dehydrogenase (PHGDH). This involves a lumping of 3 reactions 3PG + NAD+ ↔ p-hPYR + NADH+H+, p-hPYR↔pSER, pSER↔SER 𝑉𝑝ℎ𝑔𝑑ℎ 3𝑃𝐺 ∗ 𝑁𝐴𝐷 + 𝑆𝐸𝑅 ∗ 𝑁𝐴𝐷𝐻 ∗ 𝐻 + 𝑚𝑎𝑥 − 𝑉𝑟𝑝ℎ𝑔𝑑ℎ 𝐾𝑚𝑝ℎ𝑔𝑑ℎ𝑓 𝐾𝑚𝑝ℎ𝑔𝑑ℎ𝑟 = 3𝑃𝐺 𝑁𝐴𝐷 + 3𝑃𝐺 ∗ 𝑁𝐴𝐷 + 𝑆𝐸𝑅 𝑁𝐴𝐷𝐻 𝑆𝐸𝑅 ∗ 𝑁𝐴𝐷𝐻 ∗ 𝐻 + 1 + 𝑝ℎ𝑔𝑑ℎ + 𝑝ℎ𝑔𝑑ℎ + + 𝑝ℎ𝑔𝑑ℎ + 𝑝ℎ𝑔𝑑ℎ + 𝐾𝑚𝑝ℎ𝑔𝑑ℎ𝑓 𝐾 𝐾𝑚𝑝ℎ𝑔𝑑ℎ𝑟 𝐾 𝐾 𝐾 𝑚𝑎𝑥 𝑉𝑓𝑝ℎ𝑔𝑑ℎ 𝑚3𝑝𝑔 𝑚𝑛𝑎𝑑 𝑚𝑠𝑒𝑟 𝑚𝑛𝑎𝑑ℎ 21. De novo GLY synthesis through glycine hydroxymethyltransferase (GHMT, 2.1.2.1). SER↔GLY 𝑉𝑔ℎ𝑚𝑡 = 𝑆𝐸𝑅 𝐺𝐿𝑌 𝑚𝑎𝑥 −𝑉𝑟𝑔ℎ𝑚𝑡 𝐾𝑚𝑔ℎ𝑚𝑡𝑓 𝐾𝑚𝑔ℎ𝑚𝑡𝑟 𝑆𝐸𝑅 𝐺𝐿𝑌 1+ + 𝐾𝑚𝑔ℎ𝑚𝑡𝑓 𝐾𝑚𝑔ℎ𝑚𝑡𝑟 𝑚𝑎𝑥 𝑉𝑓𝑔ℎ𝑚𝑡 Parameters The parameters used in the equations and the justifications for their starting points are shown below. Steady state concentrations Starting values of model concentrations were taken from a total of several references with values below(Buxton and Frank 1997; Ercan-Fang, Gannon et al. 2002; Konig, Bulik et al. 2012). Steady State Species concentrations Species Conc., Conc range, mM mM 2.5 3.5-6.9 GLC 0.25 0.05-0.32 G6P 7.73E-02 0.01-0.1 F6P 1.55E-01 0.016-0.030 F16BP 2.00E-03 0.001-0.28 GA3P 4.14E-02 0.01-0.1 DHAP 1.00E-01 13BPG* 0.5 0.05-0.41 3PG 3.00E-02 0.007-0.05 2PG 0.15 0.012-0.27 PEP 5.00E-01 0.02-0.27 PYR 5 0.1-2.5 LAC 3.00E+00 0.5-3.5 ATP 1.18E-02 0.5-1.4 ADP 4.62E-05 0.04 AMP 4.00E+00 3.6-5.7 Pi 1.00E-03 0.03-0.05 NADH 5.49E-01 0.45 NAD+ 1.00E+01 10.0-21.0 PCr 0.04 0.026-0.034 (25mmHg) O2i** 3.00E-01 SER 1.30E-01 GLY 3.00E-01 pSER 7.00E+00 pH Extracellular concentrations 5.00E+00 GLCe 5.00E-01 LACe 3.00E-01 SERe 1.30E-01 GLYe * estimated based on Thermodynamics ** calculated based on Henry's law of solubility of O2 in water at 37°C Rate constants Starting rate constants are shown below. The Km values for substrates in glycolysis are set equal to the steady state substrate concentrations in single-substrate reactions and equal to the product of substrate concentrations in multisubstrate reactions. Km values are reported in units of mM. Vmax values are reported in units of mM/hr. Enzyme GLUT Parameter/value Vmaxtr Kmglc 100 2.1 Vmaxf Kmf 176 7.5 Vmaxf Kmf 858 0.25 Vmaxf Kmf 1769 0.23 Kmr 2.94E-10 Kmr 7.73E-02 Kmr 1.82E-10 Vmaxf 321 Vmaxf 859 Vmaxf 781 Kmf 0.16 Kmf 0.04 Kmnad 0.55 Kmr 8.29E-05 Kmr 0.002 Kmnadh 0.001 PGK Vmaxf 221 Kmr 1.5 PGM Vmaxf 527.895 Vmaxf 1340.08 9 Vmaxf 211.525 Kmf 1.18E03 Kmf 0.5 Kmf 0.03 Kmf 1.77E03 Kmnadh 0.001 Kmlac 3 Kmpyr 0.001 Kmf 3 Kmf 5 Kmf 5 Kmr 0.15 Ka 0.5 Kmnad 0.549 Kmpyr 0.5 Kmo2 0.005 Kmr 4.71E-09 Kmr 2 Kmr 2 Kmadp 0.005 HK PGI PFK ALD TPI GAPDH ENO PK LDH MCT OxPhos ATPase CKase AKase O2transp Vmaxf 434 Vmaxtr 60 Vmax 8.4 Vmaxf 390 Vmaxf 1000 Vmaxf 2000 ko2 164 Ki 0.2 Ka 0.001 Kmgap 2.00E-03 Kmpi 4 Kmbpg 1.00E-01 Kmf 4.392e-3 Kmlac 5 Kmf 5.00E-04 Kmr 2.745 Kmr 0.03 Kmr 0.15 Kmr 1.00E -11 PHGDH GHMT Other parameters Vmaxf Km3pg Kmnad Kmser 0 0.5 0.5 0.1 Vmaxf 0 nop Kmf 1.5 nfad Kmr 0.5 nnad nox 12.5 1.5 2.5 3 Kmnad h 0.01 Kmf Kmr 0.25 1.00E10 Cell uptake and release fluxes Typical uptake and release fluxes were considered(Shestov, Mancuso et al. 2013). Cell Uptake-Release Fluxes mM/h Fglc Fmpc Ftca Fldh Fltr CMRO2 Fatp(cyt) Fatp(mit) WE 80 8.8 10.7 150 150 34 150 174 14.6 Thermodynamics Standard Gibbs Free Energies (dG) were used to constrain fluxes based on Haldane relationships(Goldberg, Tewari et al. 2004; Li, Wu et al. 2011). Reaction HK PGI PFK ALD TPI GAPDH PGK PGM ENO PK LDH CK dG, kJ/mol -19.22 2.78 -15.62 24.64 7.57 2.6 -21.6 6.35 -4.47 -27.18 -23.9 -12.5 AK ATPase 0 -32.42 Supplementary References to Materials and Methods Buxton, R. B. and L. R. Frank (1997). "A model for the coupling between cerebral blood flow and oxygen metabolism during neural stimulation." J Cereb Blood Flow Metab 17(1): 64-72. Ercan-Fang, N., M. C. Gannon, et al. (2002). "Integrated effects of multiple modulators on human liver glycogen phosphorylase a." Am J Physiol Endocrinol Metab 283(1): E29-37. Goldberg, R. N., Y. B. Tewari, et al. (2004). "Thermodynamics of enzyme-catalyzed reactions--a database for quantitative biochemistry." Bioinformatics 20(16): 28742877. Konig, M., S. Bulik, et al. (2012). "Quantifying the contribution of the liver to glucose homeostasis: a detailed kinetic model of human hepatic glucose metabolism." PLoS Comput Biol 8(6): e1002577. Li, X., F. Wu, et al. (2011). "A database of thermodynamic properties of the reactions of glycolysis, the tricarboxylic acid cycle, and the pentose phosphate pathway." Database (Oxford) 2011: bar005. Shestov, A. A., A. Mancuso, et al. (2013). "Metabolic network analysis of DB1 melanoma cells: how much energy is derived from aerobic glycolysis?" Adv Exp Med Biol 765: 265-271.


![lxdfn kmf]6f]klqsf](http://s2.studylib.net/store/data/018554002_1-ad1a1817529e517aa00724ffda1d0f8d-300x300.png)