Table S1. - Springer Static Content Server
advertisement
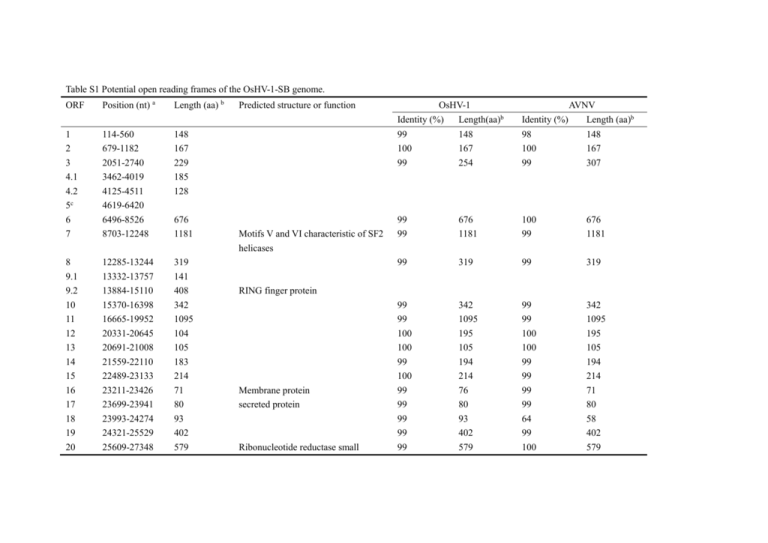
Table S1 Potential open reading frames of the OsHV-1-SB genome. ORF Position (nt) a Length (aa) b Predicted structure or function OsHV-1 AVNV Identity (%) Length(aa)b Identity (%) Length (aa)b 1 2 3 4.1 4.2 5c 6 7 114-560 679-1182 2051-2740 3462-4019 4125-4511 4619-6420 6496-8526 8703-12248 148 167 229 185 128 99 100 99 148 167 254 98 100 99 148 167 307 676 1181 99 99 676 1181 100 99 676 1181 8 9.1 9.2 10 11 12 13 14 15 16 17 18 19 20 12285-13244 13332-13757 13884-15110 15370-16398 16665-19952 20331-20645 20691-21008 21559-22110 22489-23133 23211-23426 23699-23941 23993-24274 24321-25529 25609-27348 319 141 408 342 1095 104 105 183 214 71 80 93 402 579 99 319 99 319 99 99 100 100 99 100 99 99 99 99 99 342 1095 195 105 194 214 76 80 93 402 579 99 99 100 100 99 99 99 99 64 99 100 342 1095 195 105 194 214 71 80 58 402 579 Motifs V and VI characteristic of SF2 helicases RING finger protein Membrane protein secreted protein Ribonucleotide reductase small subunit 21 22 23 24 25 26 27 28 29 30 27419-30376 30541-35439 35571-39389 39486-40619 40702-41367 41415-42602 42711-43511 43596-46157 45847-46452 46555-47301 985 1632 1272 377 221 395 266 853 201 248 31 32c 33 34 35 36 37 39 40 41 42 43 47355-47912 48173-50368 50380-51270 51350-51724 51810-52283 52353-52580 52650-53276 53521-54105 54108-55835 55985-58903 58942-60036 60155-60766 185 Primase Class I membrane protein Inactive dUTPase Related via cysteine-rich domain to ORF31 of genus Rhadinovirus and UL92 of genus Cytomegalovirus of the Herpesviridae 99 99 99 99 99 99 99 99 98 100 984 1632 1272 377 221 395 266 853 201 248 97 99 99 99 99 99 99 99 669 1632 1272 350 221 395 266 951 100 248 99 185 99 185 99 99 100 100 296 124 157 75 99 99 100 100 296 124 157 75 99 100 99 99 99 194 575 972 364 203 100 99 99 99 99 194 575 972 364 203 Encodes class I membrane protein 296 124 157 75 208 194 575 972 364 203 Inactive dUTPase Membrane protein RING finger protein BIR protein containing RING finger 125 126 127 44 45 46 47 49 60824-61771 62063-62710 62897-63712 63785-64711 64854-65579 65727-66374 66390-70628 70834-74250 315 215 271 308 241 215 1412 1138 51 75569-78076 835 52 53 54 55 56 57 78193-78735 78819-80366 80425-82848 82925-83344 83617-84465 84215-85165 180 515 807 139 282 316 58 59 60 61 62c 63c 64 65c 85211-86776 86869-90111 90164-91399 91577-93313 93472-95272 95378-97241 97281-98033 98585-100569 521 1080 411 578 250 Contains motifs V and VI characteristic of SF2 helicases Ribonucleotide reductase large subunit RING finger-like protein Class I membrane glycoprotein Multiple transmembrane protein; chloride channel Class I membrane protein RNA ligase 99 99 99 99 99 308 241 215 1412 1138 99 99 99 99 99 308 241 215 1412 1138 99 835 99 835 100 99 99 100 99 100 180 515 807 139 282 316 100 99 99 99 99 99 180 515 807 139 282 316 99 99 99 100 521 1080 411 578 99 99 99 97 521 1080 411 319 99 398 97 250 66 67 68 69 70 71 72 73c 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 100624-104016 104430-106220 106300-108390 108458-109849 109920-110933 111220-112581 112479-113045 113266-114945 115043-115399 115455-116165 116637-118673 118791-122585 122870-126094 126109-126549 126613-126963 127086-127727 127675-128565 128660-129766 129772-130128 130133-132136 132140-132547 132562-133074 133167-135413 135465-136199 136260-137144 137261-138343 1130 596 696 463 337 453 188 118 236 678 1264 1074 146 116 213 296 368 118 667 135 170 748 244 294 360 SF2 helicase Class I membrane protein Membrane protein dUTPase Class I membrane protein Membrane protein Membrane protein BIR protein lacking RING finger Class I membrane protein 99 99 99 99 82 99 100 1130 596 693 463 200 453 188 99 99 99 99 82 99 99 1130 596 694 463 200 453 188 100 100 99 99 99 100 99 100 98 99 100 99 100 99 99 99 99 100 118 236 678 1264 1151 146 116 213 296 368 118 667 135 170 748 244 294 360 100 99 99 99 99 99 99 100 98 99 99 99 99 99 99 99 99 100 118 247 678 1263 1074 146 116 213 296 368 118 667 135 170 748 244 294 360 92 93 94 95 138392-139081 139017-140231 140236-141279 141272-142297 229 404 347 341 96 97 98 99 100 101 102 103 104 105c 106 107 108 109 142363-143085 143175-143720 144223-145977 146346-147098 147589-153225 153312-153941 153982-156270 156283-157554 157702-161313 161450-163060 163205-164077 164735-166804 167020-167832 167885-170509 240 181 547 250 1878 209 762 423 1203 110 111 112 113 114 3 170604-171389 171479-172348 172472-173860 173867-174823 175046-176233 177069-177758 261 289 462 318 395 229 290 689 270 874 Similar to a family of uncharacterized, conserved eukaryotic proteins RING finger protein RING finger protein BIR protein lacking RING finger DNA polymerase Multiple transmembrane protein BIR protein containing RING finger ATPase subunit of DNA-packaging terminase Multiple transmembrane protein 99 99 99 99 229 404 347 310 99 99 99 99 229 404 347 341 100 100 99 99 99 99 99 99 99 240 181 547 250 1878 209 762 423 1203 100 99 98 99 99 99 99 99 99 240 181 547 250 1878 209 762 176 1200 97 99 99 100 465 689 270 874 97 99 99 100 464 689 270 874 99 100 99 99 43 99 261 289 462 318 494 254 99 99 99 99 43 99 261 289 462 318 494 307 2 1 116 118 119 120 121 124 122 121 120 119 118 116 178626-179129 179248-179694 180868-181638 182520-183188 183557-184315 184579-184908 186069-186716 187530-188954 190596-191750 192451-193098 194259-194588 194850-195608 195977-196645 197527-198297 a 167 148 256 222 252 109 215 474 384 215 109 252 222 256 RING finger protein RING finger protein RING finger-like protein RING finger protein indicated the positions of the ORFs in the genome the lengths of amino acid residues encoded by the ORFs c indicated disrupted genes which contain more than one small ORFs b indicated 100 99 99 98 99 99 98 97 99 98 99 99 99 99 167 148 256 222 192 76 215 474 384 215 76 192 222 256 100 98 96 98 100 98 100 97 99 100 100 100 100 97 167 148 250 222 252 109 215 474 384 215 109 252 222 250 Table S2. Primers used for genomic sequencing of OsHV-1-SB. Primer name 1F 1R 2F 2R 3F 3R 4F 4R 5F 5R 6F 6R 7F 7R 8F 8R 9F 9R 10F 10R 11F 11R 12F 12R 13F 13R 14F 14R 15F 15R 16F 16R 17F 17R 18F 18R 19F 19R 20F 20R 21F Sequence(5’-3’) TCCCGCCAATACCCATAATGCAa TGTGCCCGTTAGACTGGGAAAGa CACGCCTCTTTCCCAGTCTAACa AGGAGCCAGACCAAAACCATTC a AATTGGGATGCGATCGTTTCCTGa TGGACTTTGAGGCTCCCATTTGT a AGGAAGGCAACAGGATAGAAATG a ACAAGGAGGTTGGAAGTCGTAGAa TGGATAAATGACCGGCGAGAAAGa TGAAATCACCAACACTGGAGGCTa AAGGATCTGTCTAAGCCTCCAGTa CTCGTTGAAACATCTACCGTCAT a TTAGGTATGCCAGCCAGAGTTCAa CTTCCACCCGGAGAAAGGATTAAa TATGAACATGCCGCTTCTACTTC a CAATGCCTATAAAGACCCTGAAa TTTATCGGTGCAAATTCAGGGTCa CTAAATGGTCGCCTCAAGGTTGGa CATACAATAGTTGGGTGGTCAAa GATGATGTGGGTTCTGGTCTAAa AGGAAATTAACACCCGAGACAAa AGCACAAACACTACCGCATACAa TCTGTTCATGTATGCGGTAGTGTa ATAGCCTGCCGATATTCCTTGCa TTAGTATGGCAAGGAATATCGGa AAGGTGGTTCTGGAGTTGAGACa CAGGGTGATTGTCTCAACTCCAGa ACCCGACAGTGAACTCCCTACTCa ATGTAAATCCAAACCCTGTGCGTa GTAGTTTCATCGGTTCCAGCCTCa ATATTCTATAATCCCAGGAGCGa GATGTAACCAATGTACCACCGTa CTGAACGGTGGTACATTGGTTACa CCTATTCTCCCGGTCGTGATACAa GTATCACGACCGGGAGAATAGGa TTTACGAAACTGGGAGACCACAa CATGTTCGCTACTTGGAATGTCa AGGAGGACATCTGGCGGTA CGTGCTGAGACGGAATGTG TGTTGACTACGCCAATGACT AAGCGTCTCATCGAACTACGGGa Size (bp) 3468 3149 3874 3340 3318 3735 3222 3195 3132 3677 3680 2912 3132 3008 3533 3546 3393 3260 3584 3275 3764 21R 22F 22R 23F 23R 24F 24R 25F 25R 26F 26R 27F 27R 28F 28R 29F 29R 30F 30R 31F 31R 32F 32R 33F 33R 34F 34R 35F 35R 36F 36R 37F 37R A3 A4 39F 39R Gp3 Gp4 41F 41R 42F 42R 43F ATGTGGTCACCTTTGTGCCTGTa TGGATGTGGTTTTGGATACGAAa AGTGGTGGTGGCTCTTACTCTGa CACCGCAAATCTCCGATATGATa ATGAATTTCCAAGATAAAGATATCGGGA CCTTTCATTCACGGCATCCT TCCACTCGGAACCCCTGAGATAa ATTGCACAATATCTCAGGGGTTCa ATCCACCATAGATTGCGTCAGTa GTGAAGAAACACTGACGCAATCa AATAATCCTCGGGACCCAAGTTa ACCGGGTGACCCAGTGTTTAAa GGTGGATTCATGTGCGATCATa CTGTGATCTTGTCGAACAATTCa TTCGGTCATGTACTCTATCTCCTGa CTGTGGGATGTATAGCCGTGGAGa CCTGATTGGAGGGCAGAATGTAa GAAGTTGACTTTCTTTCCCTTGCa ACTGAGGTTGTGGGGTGTATTGTa TTTTCCCAGCAAACAGACCTATa GACATACCCAGTATTGCACCTTa TCTACACTCTGAGACCATTCCTa GATTTGTACCCAGACCAGTCATa AGGTTTCGGCGAGTCGATGAa ATGGCTTGCTGTTTTACACGTTCTa GAGACGCATTAAAATGATGGAAGa ACAGTCTGGAAATTCTCGTAGCCa CGGGCTACGAGAATTTCCAGACa CGATGATTCAGACAGGCAAGCTa ACCAACAGCTTGCCTGTCTGAAa GGAATGTCGCTTTGTACCACTGa CAGAACCTGTAATAGCAAACGAa GGGAGATGTACGACGATGTGAAa GCCAACCGTTGGAACCATAACAb GGGAATGAGGTGAACGAAACTATAGACCb TTTGTCCGAGAATTTGGTATCCa AACACCGACCATGAATCTAGGGa GGTTGTGGGTTTGGAAATGTAGAc GGCGTCCAAACTCGATTAAAc GCCGGTGAATGGATGTTATGTCa TAACCACGCTGGCTACGACTAAa CGCCTTTACCATTCATAACAAGa CCAGCATTACTGACATCCCTTAa AACCGAGGTGTAAGGGATGTCAa 3770 2139 2366 4624 3975 3131 4078 3720 3925 4456 4120 3555 3113 3731 3596 4123 1001 4369 698 3158 3050 3678 43R GATGCTCCGTGCTTGAGCCTATa 44F CCCGTTATTAGTCATCAGCTCCa 3570 a 44R TCAACGAGCAATCCCCTTTATA 45F CCGAAAATCTGATTGGCTGCTATa 2952 45R GACACCTGGGTATTGCGAGT 46F TTTGTTCACCAGACCCTTCC 3195 a 46R ATCCTTGGTGGAGTGTATAACC 47F ACAAAACTGCGGTTATACACTCa 3251 a 47R TCTTCGCCTACATATTTACTGG 48F GAAGATGAAGAGGAAGACGAAAAGa 3460 a 48R TGGGGAATACAACGTGAGATAA 49F GCTGATTAAGATTGGCAAAGAAa 2583 a 49R AACACGCTAGATATGGTCGGTA 50F TACCGACCATATCTAGCGTGTTa 3069 a 50R GGGTAACGGTTTATCATTCACA 51F AGGATGATGTGAATGATAAACCGa 2512 a 51R TGCTTTGCTCAAAACCCTCTATA 52F TTTATCTGATGCCGACAGAGTCAa 2927 a 52R TCATCTTCAAAAGCGTCGATG 53F CAGCAAAGTTTACATCGACGCTTa 3998 53R GCGCTTTCGCATTCCACATC ORF114For GGTTGGATCTCTTGGGAATGGd 2742 54R AAGATAGGCGACACGGGACT 55F TGGCAACGTCCCTATCGTAA 2191 a 55R ACTTCGTGTTGTTGTTGAGCCAT 56F GAGAAGAAGCAGCAACAGGTTAAa 2165 a 56R TTGTGGGTGTATCTGCTAGTGTC 57F ACAGGGATTACGTGTACCACAGa 2400 a 57R GCATAGGATAGCCATCAGCAAG 58F GATCCAATCACAGGCGATACCTa 2550 58R ACAGTTTGGTGGAGGAGGTG ORF121Rev ACATCCAATGAAAACAGCCGGAAd 2093 59R GGCGGGGAAAATAAGTCCTG 60F ACCACTTAACCCACAACCATATa 4449 a 60R AAGACTCCCAAGCAAGTTATGA 61F GCGCTATTGCCCATGTTAAAATa 3133 61R AGGCGATACCTTGCTGATGG 62F CCATTCATTCTCCAATCCCTC 3984 62R GGGGCGTGTCTAACCTCCTA a: Weicheng Ren. Detection methods, sequence of the complete genome of acute viral necrosis virus Isolated from Scallop, Chlamys farreri. PhD dissertation, Ocean University of China, Qingdao, China; 2009. b: Renault T, Lipart C, Arzul I. A herpes-like virus infecting Crassostrea gigas and Ruditapes philippinarum larvae in France. J Fish Dis. 2001;24(6):369-76. c : Arzul I, Nicolas JL, Davison AJ, Renault T. French scallops: A new host for Ostreid herpesvirus-1. Virology. 2001;290(2):342-9. d : Martenot C, Travaille E, Lethuillier O, Lelong C, Houssin M. Genome exploration of six variants of the Ostreid Herpesvirus 1 and characterization of large deletion in OsHV-1 mu Var specimens. Virus Res. 2013;178(2):462-70.