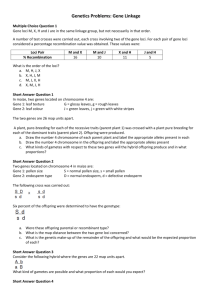
Three point linkage - Genes A, B and C: how can we tell if they are

Three point linkage - Genes A, B and C: how can we tell if they are linked?
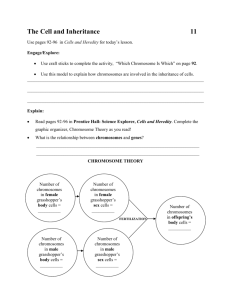
Up until now we’ve been spoiled. Genes have been assorting independently according to Mendel’s ratios and life has been easy. However, with hundreds to thousands of genes on each chromosome we are bound to run into crosses involving linked genes at some point!
The scenario: Cross AABBCC x aabbcc
• All F
1
will be AaBbCc
Testcross F
1
with aabbcc
If no linkage, expect phenotype ratio to show 1:1:1:1:1:1:1:1 ratio.
The Result:
• We get all 8 expected phenotypes, but in numbers that differ from our expected ratio. The numbers we get
are listed below.
Genotype
ABC
abc
AbC
aBc
ABc
abC
Abc
aBC
Number of observed offspring with this genotype
390
Type of gamete or chromosome (see below)
374
27
30
5
8
81
85
Total 1000
The type of chromosome/gamete column is determined by figuring out whether the combination of alleles is the same as is present in the parents OR if it is the product of recombination (crossing over during meiosis). There are 4 possibilities:
1.
Parental chromosomes – groups of alleles that are the SAME as they were in the parents. No recombination occurred with these chromosomes. Since no recombination is needed, this occurs with the greatest frequency.
Therefore, we can assume that the largest numbers of offspring inherit the unaltered parental chromosomes.
2.
Single cross over between first and second gene on the chromosome – chromosomes that are the product of crossing over between the first two loci only. (see diagram below) Frequency of this happening depends on the distance between the two loci.
3.
Single cross over between second and third gene on the chromosome– chromosomes that are the product of crossing over between the first two loci only. (see diagram below. Frequency of this happening depends on the distance between the two loci.
4.
Double cross overs – chromosomes that are the product of recombination happening between BOTH the first and second genes AND the second and third genes. Least common because TWO events need to happen, so we can always assume that the genotypes represented by the fewest offspring are the result of the double cross overs.
Let’s look at this diagrammatically -
Parental genotype: AA BB CC a a bb cc
Possible gametes o Possibility 1: pass on parental chromosomes as is with no crossing over:
A B C and a b c o Possibility 2: crossing over happens between the 1 st and 2 nd gene (effectively swapping the first gene) and a new combination is passed on.
A B C
X producing a b c a B C
A b c o Possibility 3: crossing over happens between the 2 nd and 3 rd gene (effectively swapping the last gene) and a new combination is passed on.
A B C
X producing a b c
A B c a b C o Possibility 4: crossing over happens between both pairs of genes (effectively swapping the middle gene) and a new combination is passed on.
A B C
X X producing a b c
A b C a B c
So what do we do with all this.
Step 1: Identify the parental chromosomes by identifying the two genotypes found in the highest frequency.
Parental chromosomes? _______________________ and ____________________________________
Step 2: Identify the double crossovers by identifying the two genotypes found in the lowest frequency.
Double crossovers? _____________________________and ____________________________________
Step 3: compare the double cross over combination of genes with the parental chromosomes. The locus that is different is located in the middle of the other two. Place the loci in the correct place on the chromosome below.
________-------------------------------________-------------------------------___________
Step 4: Now we look at the other options. Pick one of the pairs of single crossovers and compare it to the parental chromosomes. The locus that is different represents the site of recombination between the middle loci (identified above) and the loci that is different. Then repeat with the other pair.
Pair 1: ____________ and ______________ Which loci is different than in the parental chromosomes? _____
Pair 2: ____________ and _______________ Which loci is different than in the parental chromosomes? ______
Step 5 : Use frequencies to determine relative distance between loci
Distance between gene 1 and middle:
Add Total offspring showing single cross over between gene 1 and middle + total number of double crossovers
Divide by total number of offspring
Multiply by 100.
This equals the relative distance between the middle locus and gene 1.
Distance between the middle and gene 2:
Add Total offspring showing single cross over between gene 2 and middle + total number of double crossovers
Divide by total number of offspring
Multiply by 100.
This equals the relative distance between the middle locus and gene 2.
Draw the complete chromosome below including the proper distances between loci. ONCE you have completed the map, compare with the answer on the back to see if you are correct.
ANSWER
Genotype
ABC
abc
AbC
aBc
ABc
abC
Abc
aBC
5
8
Number of observed offspring with this genotype
390
Type of gamete or chromosome (see below)
Parental
374
27
30
Parental
Single crossover between C and B
Single crossover between C and B
Double crossover (C is locus in the middle)
81
85
Double crossover (C is locus in the middle)
Single crossover between A and C
1000
Single crossover between A and C
Total
A to C distance:
(81+85+5+8/1000) x 100 = 17.9 cM
C to B distance:
(27+30+5+8/1000) x 100 = 7.0 cM
A ______ 17.9 ____________ C _____ 7.0_____ B