mec13388-sup-0001-TableS1-S2-FigS1-S9
advertisement

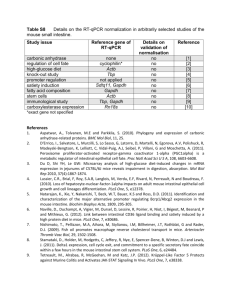
Table S1. Statistics for the de novo transcriptome assembly. Total Trinity components After Trinity assembly 66582 Total Trinity transcripts 92312 DE analysis 12003 23184 Percent GC Average contig 44.03 Median contig length 479 1079.68 Total Contig assembled N50 bases 99667695 2274 46.84 1764 2250.70 52180211 2967 Table S2. Fold change for detoxification genes that were differentially regulated in comparisons within and between populations. For within population comparisons, only genes that were induced on a population’s native host are listed. Fold changes for population differences represent values for the population in which expression was higher. CPP= coastal prickly pear; LF= laboratory food; class= EI, environmentally independent or ED, environmentally dependent; population: S= Sonora, C= Santa Catalina Island. Trinity Gene id Gene family D. moj. BLAST hit D. mel. ortholog CPP vs LF/Santa Catalina Island Saguaro Population vs differences LF/Sonora (class/population) comp39362_c1 carboxylesterase GI13369 neurotactin 3.2 2.7 comp38113_c1 carboxylesterase GI22473 GI22474 GI22476 esterase P esterase 6 6.4 comp38113_c0 carboxylesterase GI22473 GI22474 GI22476 esterase P esterase 6 comp37771_c0 carboxylesterase GI22473 GI22474 GI22476 esterase P esterase 6 18.3 comp38965_c0 carboxylesterase GI24124 α-esterase-2 11.1 comp38650_c0 carboxylesterase GI24125 neuroligin 1 3.47 comp36288_c0 carboxylesterase GI23477 α-esterase-6 comp33391_c2 carboxylesterase GI10706 CG10175 EI/C: 2.3 comp23912_c0 carboxylesterase GI13386 CG5665 EI/C: 2.9 EI/S: 27.7 EI/S: 2.3 31.4 ED/S: 4.1 4.9 comp36026_c0 carboxylesterase GI23292 CG6296 EI/C: 1.8 comp36514_c2 carboxylesterase GI15322 CG5966 3.0 6.3 comp36514_c3 carboxylesterase GI15322 CG5966 3.0 5.9 comp35387_c0 carboxylesterase GI22497 CG7367 14.4 comp38157_c0 carboxylesterase GI22654 sex-specific enzyme 2 3.0 comp34213_c0 carboxylesterase GI23290 CG6283 10.9 comp34213_c1 carboxylesterase GI23920 CG6283 11.0 comp35867_c0 carboxylesterase GI16995 CG34447 4.1 comp33195_c1 carboxylesterase GI23291 CG6295 2.4 comp36036_c0 carboxylesterase GI24275 CG8526 3.7 comp39760_c4 carboxylesterase GI23315 lipase 4 2.9 comp38330_c0 cytochrome P450 GI17433 cyp6t1 22.3 84.5 comp36625_c0 cytochrome P450 GI18694 cyp4e1 cyp4e2 cyp4e3 9.3 53.3 comp32867_c0 cytochrome P450 GI23968 GI10480 spook spookier 5.9 2.3 comp24403_c0 cytochrome P450 GI24740 cyp6d5 EI/S: 5.7 comp64131_c0 cytochrome P450 GI24740 cyp6d5 EI/S: 6.7 comp39297_c4 cytochrome P450 GI24740 cyp6d5 comp38651_c0 cytochrome P450 GI22266 cyp313a4 comp38854_c0 cytochrome P450 GI23350 GI23351 cyp304a1 comp38469_c0 cytochrome P450 GI20372 GI18650 comp31344_c0 cytochrome P450 GI17558 cyp4p3 cyp4p1 cyp4p2 cyp28a5 2.9 comp38253_c0 cytochrome P450 GI11726 cyp312a1 2.4 comp30434_c0 cytochrome P450 GI13888 Disembodied 3.9 comp38308_c1 cytochrome P450 GI11185 Phantom 3.5 comp34136_c0 glutathione S-transferase GI11974 sepia comp32794_c0 UDP-glycosyltransferase GI17522 GI17523 CG5999 CG5724 comp33150_c0 UDP-glycosyltransferase GI22630 ugt86Dd EI/S: 3.8 comp37376_c0 UDP-glycosyltransferase GI22630 ugt86Dd EI/C: 3.8 comp30485_c0 UDP-glycosyltransferase GI22630 ugt86Dd EI/S: 4.1 3.53 ED/S: 2.9 EI/S: 3.1 5.58 EI/C: 2.2 EI/C: 1.8 EI/S: 11.2 2.8 comp33973_c0 UDP-glycosyltransferase GI17058 ugt36Bb comp36246_c0 UDP-glycosyltransferase GI20954 CG17323 comp26928_c0 UDP-glucosyltransferase GI13887 CG18869 EI/C: 3.3 3.6 ED/C: 2.0 EI/C: 4.6 Figure legends Figure S1. Functional enrichment analysis for the 1,095 genes that were differentially expressed in the Santa Catalina Island population on coastal prickly pear relative to laboratory food. A false discovery rate cut-off of 0.1 was used to identify enriched terms. Node size indicates degree of significance, and larger font indicates the most significant term in cluster. Edges indicate degree of connectivity between biological terms. Figure S2. Functional enrichment analysis for the 1,541 genes that were differentially expressed in the Sonoran population on saguaro relative to laboratory food. A false discovery rate cut-off of 0.1 was used to identify enriched terms. Node size indicates degree of significance, and larger font indicates the most significant term in cluster. Edges indicate degree of connectivity between biological terms. Figure S3. Functional enrichment analysis for the 211 genes that were differentially expressed in the Santa Catalina Island population relative to the Sonoran population across all three food types (environment-independent genes). A false discovery rate cut-off of 0.1 was used to identify enriched terms. Node size indicates degree of significance, and larger font indicates the most significant term in cluster. Edges indicate degree of connectivity between biological terms. Figure S4. Functional enrichment analysis for the 32 genes that had a significant population x food interaction and were differentially expressed in the Santa Catalina Island population relative to the Sonoran population specifically on coastal prickly pear. A false discovery rate cut-off of 0.1 was used to identify enriched terms. Node size indicates degree of significance, and larger font indicates the most significant term in cluster. Edges indicate degree of connectivity between biological terms. Figure S5. Functional enrichment analysis for the 86 genes that that had a significant population x food interaction and were differentially expressed in the Sonoran population relative to the Santa Catalina Island population specifically on saguaro. A false discovery rate cut-off of 0.1 was used to identify enriched terms. Node size indicates degree of significance, and larger font indicates the most significant term in cluster. Edges indicate degree of connectivity between biological terms. Figure S6. Functional enrichment analysis for the 5,803 genes that were plastic and had overlapping reaction norms between the populations. A false discovery rate cut-off of 0.1 was used to identify enriched terms. Node size indicates degree of significance, and larger font indicates the most significant term in cluster. Edges indicate degree of connectivity between biological terms. Figure S7. Functional enrichment analysis for the 122 genes that diverged in expression between populations, but were not plastic in the Sonoran population. A false discovery rate cut-off of 0.1 was used to identify enriched terms. Node size indicates degree of significance, and larger font indicates the most significant term in cluster. Edges indicate degree of connectivity between biological terms. Figure S8. Functional enrichment analysis for the 4 genes that had reaction norms consistent with genetic assimilation. A false discovery rate cut-off of 0.1 was used to identify enriched terms. Node size indicates degree of significance, and larger font indicates the most significant term in cluster. Edges indicate degree of connectivity between biological terms. Figure S9. Functional enrichment analysis for the 65 genes that that had reaction norms consistent with the Baldwin effect. A false discovery rate cut-off of 0.1 was used to identify enriched terms. Node size indicates degree of significance, and larger font indicates the most significant term in cluster. Edges indicate degree of connectivity between biological terms. Figure S1 up-regulated down-regulated Figure S2 up-regulated down-regulated Figure S3 Figure S4 Figure S5 Figure S6 Figure S7 Figure S8 Figure S9