gcb12695-sup-0010-TableS1-S4
advertisement

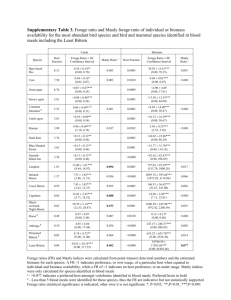
1 Supporting Information 2 Table S1 Number of individuals, collection location and coordinates, time, technique, depth and temperature, and mortalities during exposure 3 time (as % across incubation temperatures) for the prawn species investigated in this study. *Please note that for P. montagui all mortalities 4 occurred at 25 °C. Species n Locality Palaemon elegans 88 Mt Batten Palaemon macrodactylus 28 outer Thames Estuary Palaemon serratus 32 Plymouth Sound Palaemonetes varians 100 Lymington salt marsh Crangon crangon 112 Plymouth Sound 32 outer Thames Estuary Pandalus montagui Coordinates Time Technique Depth (m) Temperature (°C) June 2012 Hand held net 0.1-0.5 12.4 May 2012 Otter trawl 5-20 9.8 March 2012 Beam trawl 8-10 10.0 June 2012 Hand held net 0.3-0.5 22.3 March 2012 Beam trawl 3-10 10.0 May 2012 Otter trawl 5-20 9.8 Mortality (%) 50° 21’ 25.16’’ N 4° 07’ 35.20’’ W 8.8 51° 29’ 37.27’’ N 0° 36’ 29.27’’ W 0 50° 21’ 09.12’’ N 4° 07’ 56.09’’ W 50° 44’ 20.1’’ N 1° 32’ 16.2’’ W 0 0 50 21’ 49.28’’ N 4 11’ 13.19’’ W 0 51° 29’ 31.80’’ N 0° 50’ 53.90’’ W 25* 1 1 Table S2 GenBank accession numbers of the prawn species used for phylogenetic analysis. Species H3 12S 16S 18S 28S Palaemon elegans GU117695 DQ079729 DQ079764 DQ079807 Palaemon macrodactylus DQ642862 JQ042297 DQ642849 EF540847 EU920915 EU920938 EU920972 GQ487498 GQ487506 GQ487514 Palaemon serratus JQ042291 Palaemonetes varians JN674357 Crangon crangon EU920885 Pandalus montagui Procaris ascensionis GQ487520 GQ487495 GQ487503 GQ487511 Stenopus hispidus DQ079701 DQ079734 DQ079769 DQ079812 2 1 Table S3 Mean ± SE for oxygen consumption rate (ṀO2) and Upper Thermal Limits (UTL), measured as Loss of Orientation (LO), Onset of 2 Spasms (OS) and Death (D), for the prawn species investigated in this study after 7 d exposure to one of four incubation temperatures: 10, 15, 20 3 and 25 °C. Sample size and wet weight are also reported. *nr = not recorded Species Palaemon elegans Palaemon macrodactylus Palaemon serratus Palaemonetes varians Crangon crangon Temperature (°C) n ṀO2 (nmol O2 g wet weight -1 min-1) LO (°C) OS (°C) D (°C) Wet weight (g) Wet weight range (g) 10 15 20 25 10 15 20 25 10 15 20 25 10 15 20 25 10 15 20 25 18 20 17 16 5 7 7 7 8 8 8 7 18 17 16 10 27 23 20 21 90.7 ± 8.4 249.2 ± 14.1 319.3 ± 20.9 699.7 ± 76.9 65.3 ± 15.1 80.9 ± 4.9 157.5 ± 10.0 304.5 ± 21.8 46.1 ± 7.1 70.2 ± 8.4 147.4 ± 21.8 171.4 ± 7.6 80.1 ± 7.7 195.3 ± 13.8 434.6 ± 50.6 799.9 ± 56.7 48.0 ± 3.1 108.3 ± 8.4 134.4 ± 13.0 215.0 ± 16.4 29.7± 0.3 31.2 ± 0.2 33.3 ± 0.3 34.5 ± 0.4 29.7 ± 0.6 32.4 ± 0.3 33.4 ± 0.5 35.7 ± 0.4 28.2 ± 1.1 30.9 ± 0.4 32.2 ± 0.3 31.6 ± 0.6 30.1 ± 0.5 31.8 ± 0.3 32.6 ± 0.2 34.1 ± 0.2 30.7 ± 0.2 32.2 ± 0.2 34.1 ± 0.2 34.3 ± 0.2 30.6 ± 0.3 32.9 ± 0.2 34.5 ± 0.3 35.0 ± 0.4 31.8 ± 0.5 33.7 ± 0.6 35.3 ± 0.2 37.8 ± 0.5 28.6 ± 0.5 32.0 ± 0.4 33.0 ± 0.8 33.5 ± 0.8 32.3 ± 0.1 33.7 ± 0.2 34.1 ± 0.1 36.0 ± 0.1 31.7 ± 0.2 33.3 ± 0.2 35.4 ± 0.2 34.4 ± 0.5 33.8 ± 0.2 35.5 ± 0.2 37.2 ± 0.3 37.9 ± 0.2 33.6 ± 0.6 35.9 ± 0.3 37.3 ± 0.1 39.9 ± 0.2 30.9 ± 0.5 33.4 ± 0.2 36.5 ± 0.2 37.6 ± 0.1 36.1 ± 0.3 36.7 ± 0.2 35.9 ± 0.1 37.3 ± 0.1 33.7 ± 0.1 35.7 ± 0.1 36.7 ± 0.1 37.4 ± 0.1 0.3 ± 0.0 0.3 ± 0.0 0.4 ± 0.0 0.5 ± 0.1 0.9 ± 0.1 0.7 ± 0.1 0.8 ± 0.1 0.7 ± 0.1 1.5 ± 0.3 1.1 ± 0.1 1.0 ± 0.1 1.2 ± 0.1 0.3 ± 0.0 0.3 ± 0.0 0.3 ± 0.0 0.3 ± 0.0 0.6 ± 0.0 0.8 ± 0.1 0.8 ± 0.1 0.8 ± 0.1 0.18 – 0.62 0.14 – 0.74 0.16 – 0.86 0.16 – 1.13 0.61 – 1.10 0.39 – 1.03 0.48 – 0.97 0.43 – 1.10 0.73 – 2.91 0.87 – 1.50 0.56 – 1.56 0.88 – 1.82 0.16 – 0.36 0.18 – 0.65 0.14 – 0.49 0.19 – 0.39 0.32 – 1.29 0.26 – 1.48 0.21 – 1.55 0.32 – 1.44 3 Pandalus montagui 10 15 20 25 8 8 5 8 96.6 ± 3.1 105.9 ± 7.8 193.9 ± 4.4 nr* 23.6 ± 0.2 25.2 ± 1.0 26.0 ± 1.0 nr* 24.8 ± 0.3 27.4 ± 0.2 27.6 ± 0.7 nr* 27.1 ± 0.4 30.5 ± 0.1 30.5 ± 0.3 nr* 1.3 ± 0.1 1.2 ± 0.1 1.1 ± 0.1 nr* 0.81 – 1.85 0.87 – 1.46 0.73 – 1.48 nr* 4 1 Table S4 Mean plasticity of oxygen consumption rate (ΔṀO2) and Upper Thermal Limits 2 (ΔUTL), measured as LO, OS and D, calculated at five temperature intervals (10-15, 15-20, 3 20-25, 10-25 and 10-20 °C) as the difference between mean values of ṀO2 and UTL either 4 between consecutive or extreme incubation temperatures (considering either 25 or 20 °C as 5 upper extreme temperature). nr* = not recorded Species Palaemon elegans Palaemon macrodactylus Palaemon serratus Palaemonetes varians Crangon crangon Pandalus montagui Temperature (°C) ΔṀO2 (nmol O2 g wet weight -1 min-1) ΔUTL LO (°C) ΔUTL OS (°C) ΔUTL D (°C) 10-15 15-20 20-25 10-25 10-20 10-15 15-20 20-25 10-25 10-20 10-15 15-20 20-25 10-25 10-20 10-15 15-20 20-25 10-25 10-20 10-15 15-20 20-25 10-25 10-20 158.5 70.1 380.4 609.0 228.6 15.6 76.6 147.0 239.2 92.2 24.2 77.2 24.0 125.3 101.4 115.2 239.3 365.3 719.8 354.5 60.1 26.3 80.6 167.0 86.4 1.4 2.1 1.4 4.8 3.5 2.7 1.0 2.3 6.0 3.7 2.7 1.4 -0.7 3.4 4.1 1.7 0.8 1.5 4.0 2.5 1.5 1.9 0.2 3.7 3.4 2.3 1.6 0.5 4.4 3.9 1.8 1.6 2.5 5.9 3.4 3.4 1.0 0.4 4.9 4.4 1.5 0.4 1.9 3.8 1.8 1.6 2.1 -1.0 2.7 3.7 1.6 1.8 0.6 4.0 3.4 2.4 1.4 2.6 6.4 3.8 2.6 3.1 1.1 6.8 5.7 0.6 -0.8 1.4 1.2 -0.1 2.1 1.0 0.7 3.7 3.0 10-15 15-20 20-25 10-25 10-20 9.3 88.0 nr* nr* 97.3 1.6 0.8 nr* nr* 2.4 3.0 -0.2 nr* nr* 2.8 3.4 0.0 nr* nr* 3.4 5 1 Figure S1 Representation of plasticity, here calculated according to the concept of Reaction 2 Norms (Pigliucci, 2001; Schlichting & Pigliucci, 1998). In order to identify both the 3 magnitude and shape of Reaction Norms (sensu Pigliucci, 2001) plasticity is calculated both 4 within the whole temperature range examined in this study (P10-25) (red arrow) and within 5 smaller temperature intervals (P10-15, P15-20, P20-25) (blue arrows). In the first case, plasticity is 6 calculated as the difference between mean values of either oxygen consumption rate (ṀO2) or 7 Upper Thermal Limits (UTL) measured at the two extreme incubation temperatures tested, 8 while in the second case it is calculated as the difference between either mean ṀO2 or mean 9 UTL measured at two consecutive incubation temperatures. 10 11 Figure S2 Bayesian majority-rule consensus phylogram of the combined 12S rDNA, 16S 12 rDNA, 18S rDNA and 28S rDNA and H3 data set. Bootstrap values (1000 replicates) for 13 nodes obtained by parsimony analysis are indicated by numbers placed above the nodes. Only 14 values equal to or greater than 60 are shown. Types of habitat are indicated by letters placed 15 next to species names: B stands for broader/more stable thermal niches, while N indicates 16 species characterised by narrower/more stable thermal niches. ‘*’ indicates that, whilst P. 17 macrodactylus can occupy broad thermal niches, this species actually occurs in subtidal 18 habitats. However, for the purpose of our work, this species is classed among those occupying 19 broader/more variable thermal niches. 20 21 Figure S3 The effect of temperature on UTL, measured as (a) Loss of Orientation (LO) and (b) 22 Death (D), and (c) UTL plasticity in the prawn species investigated in this study: Palaemon 23 elegans, Palaemon macrodactylus, Palaemon serratus, Palaemonetes varians, Crangon 24 crangon and Pandalus montagui. Histograms represent mean ± SE for UTL after 7 d exposure 25 to one of four incubation temperatures: 10 (yellow), 15 (light orange), 20 (dark orange) and 6 1 25 °C (red). Significantly different mean UTL (p < 0.05) at different incubation temperatures 2 for the same species are indicated by different letters placed above the histograms, while 3 significantly different mean UTL (p < 0.05) at the same incubation temperature among 4 different species are indicated by different numbers placed inside the histograms. Finally, 5 overall significant differences in mean UTL (p < 0.05) among different species are indicated 6 by different numbers preceded by a star placed at the top of the graph above each species. 7 Pairwise comparisons were conducted using the Estimated Marginal Means test with Least 8 Significant Difference test correction. Lines in (c) represent Upper Thermal Limits– 9 Temperature (UTL–T): i.e. the patterns through which species increased their UTL in 10 response to increasing incubation temperature according to the best-fit regression model. UTL 11 are measured as LO (grey) and D (black). Raw data, regression equation and relevant 12 statistics for UTL are provided in Figure S5. 13 14 Figure S4 The relationship between temperature and ṀO2 for the prawn species investigated 15 in this study. Circles represent individual prawn ṀO2 measured after 7 d exposure to one of 16 four incubation temperatures: 10 (yellow), 15 (light orange), 20 (dark orange) and 25 °C (red). 17 Full lines represent the best-fit significant regression model; regression equation and relevant 18 statistics are as follows: P. elegans: y = 28.037e0.126x, R² = 0.784, F1,69 = 249.8, p < 0.0001; P. 19 macrodactylus: y = 17.024e0.112x, R² = 0.856, F2,23 = 142.7, p < 0.0001; P. serratus: y = 20 16.375e0.098x, R² = 0.743, F1,29 = 83.6, p < 0.0001; P. varians: y = 16.242e0.158x, R² = 0.839, 21 F1,58 = 301.9, P < 0.0001; C. crangon: y = 19.601e0.095x, R² = 0.672, F1,89 = 182.0, p < 0.0001; 22 P. montagui: y = 1.574x2 - 37.499x + 314.214, R² = 0.896, F2,17 = 73.5, p < 0.0001. 23 24 Figure S5 The relationship between temperature and UTL, measured as (a) LO, (b) OS and (c) 25 D, for the prawn species investigated in this study. Circles represent individual prawn UTL 7 1 measured after 7 d exposure to one of four incubation temperatures: 10 (yellow), 15 (light 2 orange), 20 (dark orange) and 25 °C (red). Full and dotted lines represent the best-fit 3 significant and marginally significant regression model respectively; regression equation and 4 relevant statistics are as follows: (a) P. elegans: y = 0.330x + 26.385, R² = 0.739, F1,64 = 180.8, 5 p < 0.0001; P. macrodactylus: y = 0.375x + 26.306, R² = 0.798, F1,24 = 95.0, p < 0.0001; P. 6 serratus: y = 4.095ln(x) + 19.275, R² = 0.436, F1,25 = 19.3, p < 0.0001; P. varians: y = 0.255x 7 + 27.689, R² = 0.544, F1,53 = 63.2, p < 0.0001; C. crangon: y = 4.285ln(x) + 20.791, R² = 8 0.657, F1,84 = 160.8, p < 0.0001; P. montgui: y = 3.468ln(x) + 15.685, R² = 0.217 F1,13 = 3.6, p 9 = 0.080; (b) P. elegans: y = 4.981ln(x) + 19.286, R² = 0.655, F1,61 = 115.8, p < 0.0001; P. 10 macrodactylus: y = 0.390x + 27.775, R² = 0.905, F1,21 = 199.4, p < 0.0001; P. serratus: y = 11 5.335ln(x) + 16.775, R² = 0.595, F1,23 = 33.8, p < 0.0001; P. varians: y = 0.226x + 30.041, R² 12 = 0.808, F 1,53 = 222.9, p < 0.0001; C. crangon: y = 3.575ln(x) + 23.724, R² = 0.410, F1,77 = 13 53.6, p < 0.0001; P. montgui: y = 2.233x2 - 0.065x + 8.975, R² = 0.814, F2,15 = 32.9, p < 14 0.0001; (c) P. elegans: y = 4.564ln(x) + 23.284, R² = 0.734, F1,68 = 187.7, p < 0.0001; P. 15 macrodactylus: y = 0.406x + 29.580, R² = 0.919, F1,23 = 260.9, p < 0.0001; P. serratus: y = 16 7.730ln(x) + 12.922, R² = 0.920, F1,29 = 335.0, p < 0.0001; P. varians: y = 0.050x + 35.618, 17 R² = 0.079, F1,53 = 4.5, p = 0.038; C. crangon: y = 4.100ln(x) + 24.354, R² = 0.903, F1,87 = 18 805.9, p < 0.0001; P. montgui: y = -0.069x2 + 2.415x + 9.860, R² = 0.962, F 1,18= 216.1, p < 19 0.0001. 20 21 Figure S6 The relationship between ṀO2 and UTL, measured as (a) LO and (b) D, for the 22 prawn species investigated in this study. Circles represent individual prawn ṀO2 and UTL 23 measured after 7 d exposure to one of four incubation temperatures: 10 (yellow), 15 (light 24 orange), 20 (dark orange) and 25 °C (red). Full and dotted lines represent the best-fit 25 significant and marginally significant regression models respectively; regression equation and 8 1 relevant statistics are as follows: (a) P. elegans: y = 2.243ln(x) + 19.689, R² = 0.677, F1,64 = 2 134.4, p < 0.0001; P. macrodactylus: y = 3.108ln(x) + 17.963, R² = 0.808, F1,24 = 101.2, p < 3 0.0001; P. serratus: y = 1.792ln(x) + 22.777, R² = 0.294, F1,25 = 10.4, p = 0.003; P. varians: y 4 = 1.369ln(x) + 24.478, R² = 0.486, F1,53 = 50.2, p < 0.0001; C. crangon: y = 1.759ln(x) + 5 24.608, R² = 0.383, F1,84 = 52.1, p < 0.0001; P. montgui: y = 2.790ln(x) + 11.619, R² = 0.219, 6 F1,13 = 3.7, p = 0.078; (b) P. elegans: y = 1.836ln(x) + 25.908, R² = 0.640, F1,68 = 121.0, p < 7 0.0001; P. macrodactylus: y = 3.332ln(x) + 20.669, R² = 0.834, F1,23 = 115.6, p < 0.0001; P. 8 serratus: y = 3.491ln(x) + 18.839, R² = 0.641, F1,29 = 51.8, p < 0.0001; P. varians: y = 9 0.306ln(x) + 34.780, R² = 0.093, F1,53 = 5.4, p = 0.023; C. crangon: y = 1.856ln(x) + 27.194, 10 R² = 0.642, F1,87 = 156.1, p < 0.0001; P. montgui: p > 0.05. 9