JCP-Supplementary material_R2
advertisement

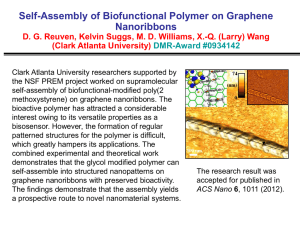
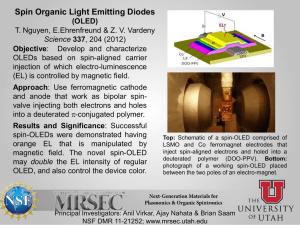
Supplementary material Molecular simulation study of role of polymer–particle interactions in the straindependent viscoelasticity of elastomers (Payne effect) Yulong Chen,2,3 Ziwei Li,2,3 Shipeng Wen,1,3 Qingyuan Yang,1,5,a) Liqun Zhang, Chongli Zhong,1,5 and Li Liu2,3,4,a) 1State Key Laboratory of Organic-Inorganic Composites, Beijing University of Chemical Technology, Beijing 100029, China 2State Key Laboratory of Chemical Resource Engineering, Beijing University of Chemical Technology, Beijing 100029, China 3Beijing Engineering Research Center of Advanced Elastomers, Beijing University of Chemical Technology, Beijing 100029, China 4Key Laboratory of Carbon Fiber and Functional Polymers, Ministry of Education, Beijing University of Chemical Technology, Beijing 100029, China 5Laboratory of Advanced Nanostructural Materials, Beijing University of Chemical Technology, Beijing 100029, China a)Corresponding author: qyyang@mail.buct.edu.cn, LiuL@mail.buct.edu.cn This supplementary document contains the following material. SI. Details of the initial simulation system construction SII. Mean-square displacements of NPs across polymer network after crosslinking SIII. Additional analyses of polymer dynamics and the calculations of mean relaxation times (MRT) of polymer monomers. SIV. Interfacial potential energy of polymers from NPs during oscillatory shear SV. Comparison of the IIDS factors of polymer in unfilled systems at different temperatures and monomers located in first layer in filled systems SVI. Strain-dependent viscoelasticity of filled system at different temperatures 1 SI. Details of the initial simulation system construction To generate the initial configuration for each simulation system, all the polymer chains, crosslinking agent molecules (CLAs) and nanoparticles (NPs) were placed in a very large simulation box, so the densities of the systems were low (the number densities of the polymer beads (PBs) were about * = 0.01). Such systems were first relaxed using equilibrium molecular dynamics (EMD) simulation in the canonical (NVT) ensemble at the fixed temperature T* = 1.0 without any attractive interactions for 2103 (2106 timesteps). Then, all the attractions involved were switched on (the polymer–particle interaction strength (pn) was set at 2.0 so that the NPs could be well dispersed in the matrix before the crosslinking procedure1) and the isothermal-isobaric (NPT) ensemble was performed at temperature T* = 1.0 and pressure P* = 0 to reduce the system volume and to increase the number density of PBs to the melt density (around * = 0.85). Following this, all structures of the systems were equilibrated over time. To make sure whether the systems reached equilibrium, the mean-square radius of gyration 𝑅𝑔2 of the polymer chains with time have been monitored (see Figure S1(a)). It was found that 4𝑅𝑔2 fluctuated around 29.4 (𝑅𝑔 =2.71) after 2.5103. This is the signature when system reached equilibrium. At the same time, the mean-square displacement (MSD) of molecules exceeded 4𝑅𝑔2 at 2.5103 (see Figure S1(b)), meaning that the polymer chains moved by at last 2𝑅𝑔 when the systems equilibrated. In addition, the dispersion states of NPs, using snapshots and radial distribution function, were also checked to confirm the equilibrium of the systems. In the current study, all the systems were implemented for at least 5103 to ensure fully equilibrated. (a) MSD of molecules 32 4Rg 2 31 30 29 28 27 0 3 1x10 3 2x10 3 3x10 time/ 3 4x10 3 5x10 60 (b) 50 40 30 20 10 0 0 3 1x10 3 2x10 3 3x10 time/ 3 4x10 3 5x10 FIG. S1. (a) Running values of the mean-square radius of gyration 𝑅𝑔2 with simulation time, (b) mean-square displacement (MSD) of polymer chains, i.e., MSD of center of mass of polymer chains, as a function of time, for the system with NP volume fraction = 0.152 and polymer–particle interaction εpn = 2.0. 2 Once reaching equilibrium, chemical bonds between PBs and CLAs were generated if their distances were less than 1.25, where one CLA was allowed to link two PBs with only one CLA for each PB. These bonds were also described using the FENE potential with parameters the same as those for the bonds in the polymer. Through this procedure, all the polymer chains could be connected by the CLAs to form the crosslinked network. Then, pn were tuned in a broad range (from 4.0 to 12.0) and NPT-MD simulation at T* = 1.0 and P* = 0 was further conducted to equilibrate each system for about 104 until no evident change was achieved for the results with longer time. Depending on the filler loading and polymer–particle interaction, the number densities of PBs obtained were about * = 0.86–0.92, and the number densities of crosslinking sites were found to range from about 0.04 to 0.05 –3, which were defined as ρcl = Ncl/V, where Ncl is the number of the CLAs (Ncl = 800) and V is volume of simulation box (ranging from 15 000 to 20 000 3). It has been demonstrated that when mapping the coarse-grained bead–spring model to real polymers, the diameter () of the PB corresponded to segmental lengths of about 0.5 to 1.3 nm,2 depending on the specific polymers. As a result, the crosslinking densities of the simulation systems were approximately 3 10 5 to 7 10 4 mol/cm3, which were within the ranges (10–5 to 10–4 mol/cm3) for real elastomers.3 It should be noted that there were a certain number of cross-links between the monomers in the same chain (self-linkage). In fact, the formation of self-linkages is certainly possible in real crosslinking or vulcanization processes. 3 SII. Mean-square displacements of NPs across polymer network after crosslinking It is generally accepted that crosslinking can prevent the diffusion of NPs to some extent. In order to check whether the NPs could truly diffuse across the polymer network, after crosslinking, we have measured the MSD of NPs with time, as shown in Figure S2. The slope of the MSD versus time is proportional to the diffusion rate of the NPs. It can be found that in the initial very short simulation time, the diffusion rate increased with increasing polymer–particle interaction, which was due to the thermodynamically non-equilibrium states of the systems when the polymer–particle interaction has just been tuned from 2.0 to larger value at the beginning of this stage. In the following stage, as the systems gradually reached equilibrium, the diffusion rate of the NPs decreased. Finally, the higher the εpn was, the lower the diffusion rate behaved, because the polymer segments were tightly bound to the NPs at high polymer–particle attraction that can hinder the diffusion of the NPs. These results demonstrated that the NPs can diffuse in the network of our model systems. MSD of NPs 10 pn = 4.0 8 pn = 8.0 6 pn = 12.0 4 2 0 0 3 1x10 3 2x10 3 3x10 3 4x10 3 5x10 time/ FIG. S2. Mean-square displacement (MSD) of nanoparticles (NPs) as a function of time for systems ( = 0.152) after cross-linked. 4 SIII. Additional analyses of polymer dynamics and the calculations of mean relaxation times (MRT) of polymer monomers. A dynamic parameter, i.e., the incoherent intermediate dynamic structure (IIDS) factor qs (t ) , was adopted to characterize the dynamics of polymers at the segmental scale in the initial systems, as defined by4-7 qs (t ) 1 M 1 M sin(qrm (t )) exp[ iq (rm (t ) rm (0))] M m1 M m1 qrm (t ) (1) where M is the number of polymer monomers; q and q are the wave vector and number, respectively; (rm(t) – rm(0)) and rm (t ) are the displacement vector and scalar of monomer m after time t, respectively. Similar to the literature,7 the value of q was set to 6.9 in this study which also corresponds to the first peak location of the static structure factor obtained from our unfilled system (Figure S3). Figure S4 shows the IIDS factors calculated for polymer monomers located in the first layer that surrounding the NPs (one bead thickness) and that of all the monomers in the systems with three different filler volume fractions, where the result for the unfilled system is also included for comparison. It can be found that within the broad range of pn examined, the IIDS factors for the monomers in the first layer almost coincide for all the systems if t ≤ 0.1. This short-time regime corresponds to the ballistic motion of a monomer. 6 As time increased, the decay of qs (t ) becomes much slower with increasing polymer–particle interaction. This can be explained by the fact that higher interfacial strength will result in a more significant reduction of the mobility of chain segments near the NP surfaces.8,9 In addition, the increase of filler volume fraction can also slow down the decay of qs (t ) , since the monomer feel more constraints imposed by its neighboring NPs. The decay of qs (t ) for all the monomers in each filled system also manifests a similar behavior to that of the monomers in the first layer but with a faster rate, and both of them are slower than that for the neat system, as can be reflected from the comparisons given in Figures S4. On the other hand, for the systems with weak polymer– particle interactions and low filler loadings, the changes of qs (t ) for all the monomers are very limited (dash lines, Figure S4(a), while the decay rates become significantly slower with increasing εpn or , and even can be close to those for the monomers located in the first layer (Figure S4(c)) because of more monomers subjected to the constraints from NPs. Moreover, for the systems with = 0.265 and εpn = 8.0–12.0, the decay curves of qs (t ) 5 occur a plateau after t > 0.1 and then drop quickly, as shown in Figure S4(c). The former phenomenon is generally regarded as the β-relaxation with the latter as the α-relaxation.6 In the β-relaxation regime, the monomers move within the cages composed by their nearest neighbors (monomers or NPs), while in the αrelaxation regime, the monomers start to leave these cages, and the temporarily frozen structures “melts”. In addition, the mean relaxation time (MRT), i.e., the α-relaxation time τα, of all the polymer monomers and the monomers present in the first layer (one bead thickness) in each system was calculated which was defined as the time where qs (t ) decays to 1/e.7 6 2.5 S (q ) 2.0 1.5 1.0 0.5 0.0 4 8 12 16 q 20 FIG. S3. Static structure factor S(q) for the neat elastomeric system, which was calculated according to the equation given in ref. 7. The location of the first peak corresponds to q = 6.9. 1.0 1.0 (a) 0.8 0.6 s s q (t) q (t) pn Increasing 0.6 0.4 0.2 0.0 -2 10 (b) 0.8 0.4 0.2 -1 10 0 10 t/ 1 0.0 -2 10 2 10 10 -1 0 10 10 1 t/ 10 2 10 3 10 1.0 (c) -rela 0.8 xation - re l ax s q (t) 0.6 ati on 0.4 0.2 0.0 -2 10 -1 10 0 10 1 10 t/ 2 10 3 10 4 10 FIG. S4. Incoherent intermediate dynamic structure functions ( qs (t ) ) for the polymer monomers located in the first layer (solid lines) and for all the monomers (dash lines) in the systems with different polymer–particle interactions (pn ranges from 4.0 to 12.0) at three filler volume fractions: (a) = 0.066, (b) = 0.152, and (c) = 0.265. In each subfigure, the result for the unfilled system is also included for comparison (black dash line). 7 SIV. Interfacial potential energy of polymers from NPs during oscillatory shear -24100 (a) -24200 Eint -24300 -24400 -24500 -24600 -24700 0 -24100 50 100 t/ 150 200 50 100 150 200 (b) -24200 Eint -24300 -24400 -24500 -24600 -24700 0 t/ FIG. S5. Interfacial potential energy Eint (in units of ε) of polymers from NPs in the system with = 0.066 and pn = 10.0 during two periods of oscillatory shear at strain amplitudes of (a) γ0 = 0.1 and (b) γ0 = 0.4 with shear frequency = 0.01. 8 SV. Comparison of the IIDS factors of polymer in unfilled systems at different temperatures and monomers located in first layer in filled systems 1.15 1.10 V(T) 1.05 1.00 Tg 0.95 0.90 0.0 0.2 0.4 0.6 0.8 1.0 T FIG. S6. Dependence of the effective volume V(T) per monomer on temperature for the unfilled cross-linked system. The abrupt change in the curve is generally considered as a signature of the glass transition for polymers. The results indicate that the glass transition temperature Tg was about 0.4. 1.0 T=0.1 T=0.2 T=0.3 T=0.4 0.6 s q(t) 0.8 0.4 T=0.5 T=0.6 T=0.8 0.2 First layer T=1.0 0.0 -2 10 -1 10 0 10 1 10 t/ 2 10 3 10 4 10 FIG. S7. Influence of temperature on the incoherent intermediate dynamic structure factor qs (t ) for all the monomers in the unfilled system. The result for the monomers located in the first layer (black line) around nanoparticles in the filled system is also shown for comparison, where the filler volume fraction was = 0.265 and the polymer–particle interaction was pn = 12.0. It can be seen that the relaxation time for the monomers in the first layer is faster than that for the unfilled system at temperatures below Tg, indicating that the polymer shell in such system still not in the glassy state. 9 SVI. Strain-dependent viscoelasticity of filled system at different temperatures 20 5 G' G" 12 6 (a) T = 0.6 T = 0.8 T = 1.0 16 3 4 2 0.1 0.2 0.3 0 0.4 0.5 (b) 0.4 0.5 4 8 0 0.0 T = 0.6 T = 0.8 T = 1.0 1 0.0 0.1 0.2 0.3 0 FIG. S8. Dependence of (a) storage modulus G′ and (b) loss modulus G″ on the shear strain amplitude for the filled system with = 0.152 and with high polymer–particle interaction strength (pn = 10.0) at three different temperatures, where the shear frequency = 0.01. References 1 J. Liu, Y. Gao, D. Cao, L. Zhang, and Z. Guo, Langmuir 27, 7926 (2011). 2 K. Kremer and G. S. Grest, J. Chem. Phys. 92, 5057 (1990). 3 A. Vieyres, R. Pérez-Aparicio, P.-A. Albouy, O. Sanseau, K. Saalwächter, D. R. Long, and P. Sotta, Macromolecules, 46, 889 (2013). 4 C. Bennemann, W. Paul, J. Baschnagel, and K. Binder, J. Phys.: Condens. Matter 11, 2179 (1999). 5 C. Bennemann, W. Paul, K. Binder, and B. Dünweg, Phys. Rev. E 57, 843 (1998). 6 C. Bennemann, J. Baschnagel, and W. Paul, Eur. Phys. J. B 10, 323 (1999). 7 J. Liu, D. Cao, L. Zhang, and W. Wang, Macromolecules 42, 2831 (2009). 8 G. D. Smith, D. Bedrov, and O. Borodin, Phys. Rev. Lett. 90, 226103 (2003). 9 Y. Pu, M. H. Rafailovich, J. Sokolov, D. Gersappe, T. Peterson, W.-L. Wu, and S. A. Schwarz, Phys. Rev. Lett. 87, 206101 (2001). 10