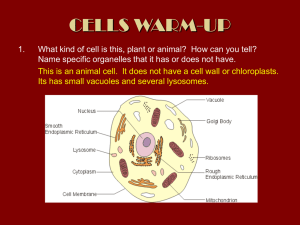
cells
advertisement

BERNARD et al SELECT MICROTUBULE INHIBITORS DISRUPT LYSOSOMES SUPPLEMENTARY INFORMATION Select microtubule inhibitors increase lysosome acidity and promote lysosomal disruption in acute myeloid leukemia (AML) cells Dannie Bernard1, Marinella Gebbia2, Swayam Prabha1, Marcela Gronda1, Neil MacLean1, Xiaoming Wang1, Rose Hurren1, Mahadeo A. Sukhai1, Eunice E. Cho1, Morris F. Manolson3, Alessandro Datti4,5, Jeffrey Wrana4, Mark D. Minden1, Rima Al-Awar6, Ahmed Aman6, Corey Nislow7, Guri Giaever7 and Aaron D. Schimmer1 AFFILIATIONS: 1Princess Margaret Cancer Centre, University Health Network, Toronto, ON, Canada; 2Department of Molecular Genetics, Donnelly Centre for Cellular and Biomolecular Research, University of Toronto, Toronto, ON, Canada; 3Faculty of Dentistry, Dental Research Institute, University of Toronto, Toronto, ON, Canada; 4Samuel Lunenfeld Research Institute, Mount Sinai Hospital, Toronto, ON, Canada; 5 Department of Agricultural, Food, and Environmental Sciences, University of Perugia, Perugia, Italy ; 6Drug Discovery Program, Ontario Institute for Cancer Research, Toronto, ON, Canada and 7Faculty of Pharmaceutical Sciences, University of British Columbia, Vancouver, BC, Canada CORRESPONDING AUTHOR: Aaron D. Schimmer, Princess Margaret Hospital, Rm 9-516, 610 University Ave, Toronto, Ontario, Canada M5G 2M9. Phone: 416.946.2838; Fax: 416.946.6546; E-mail: aaron.schimmer@utoronto.ca. 1 BERNARD et al SELECT MICROTUBULE INHIBITORS DISRUPT LYSOSOMES SUPPLEMENTARY MATERIALS AND METHODS Drugs and reagents The Natural Product chemical library was purchased from MicroSource Discovery Systems (Gaylordsville, CT). Deoxysappanone B 7,4’ dimethyl ether (Deox B 7,4) and its reduced form (Red-Deox B 7,4) were synthesized at the Ontario Institute for Cancer Research (Toronto, ON, Canada; see supplemental information for synthesis method). The enantiomer separation of Deox B 7,4 and Red-Deox B 7,4 was outsourced (Lotus Separations, Princeton, NJ). Nocodazole was obtained from Adipogen (San Diego, CA), vincristine was supplied by Abcam (Cambridge, MA) and other microtubule inhibitors as well as adriamycin were purchased from Sigma Aldrich (Oakville, ON, Canada). Bafilomycin A1 was acquired from Cayman Chemicals (Ann Arbor, MI). All drugs were prepared in dimethyl sulfoxide (DMSO). Unless otherwise stated, all other chemicals were purchased from Sigma Aldrich (Oakville, ON, Canada). Cell culture The leukemia cell lines TEX, OCI-AML2, K562 and THP1 were maintained in Iscove’s modified Dulbecco’s Medium (IMDM). maintained in RPMI 1640 medium. The leukemia cell lines HL60 and U937 were The ovarian adenocarcinoma cell lines A2780 and A2780ADR[1], were provided by Dr. Jeremy Squire (Kingston General Hospital, Kingston, ON, Canada) and were also maintained in RPMI 1640 medium. A2780ADR cells were treated overnight once a week with 0.1μg/ml adriamycin to maintain the drug resistance phenotype. The epidermoid carcinoma cell lines KB-3-1 and KB-4.0-HTI36 (a gift from Dr. F. Loganzo, Pearl River, NY)[2] were grown in Dulbecco’s Modified Eagle Medium (DMEM). For all cell lines with the exception of TEX, medium was supplemented with 10% fetal bovine serum (FBS), 2 BERNARD et al SELECT MICROTUBULE INHIBITORS DISRUPT LYSOSOMES 100μg/ml penicillin and 100 U/ml of streptomycin (all from Hyclone, Logan, UT). TEX cells were grown in the presence of 15% FBS, 100μg/ml penicillin, 100 U/ml of streptomycin, 2mM L-glutamine, 20ng/ml SCF, 2ng/ml IL-3. All cells were incubated at 37°C in a humidified air atmosphere supplemented with 5% CO2. For hypoxia experiments, cells were transferred to hypoxic culture chambers (MACS VA500 microaerophilic workstation, H35 HypoxyWorkStation; Don Whitley Scientific) in which the atmosphere consisted of 5% H2, 5% CO2 and either 0.2%, 1% or 3% O2 as well as residual N2. Colony formation assay The collection and use of human tissue for this study was approved by the local ethics review board (University Health Network, Toronto, ON, Canada). Peripheral blood samples from primary AML patients and GM-CSF-mobilized peripheral blood stem cells (PBSCs) from volunteers donating PBSCs for allotransplant were obtained after informed consent. Blood samples were subjected to light density isolation with Ficoll-Paque Plus (GE Healthcare, Uppsala, Sweden) to separate mononuclear cells (MNCs). MNCs (1x105/dish) were plated in 0.1ml IMDM supplemented with 10% FBS and 0.9ml MethoCult GF H4434 medium (StemCell Technologies, Vancouver, BC, Canada) in duplicate 35mm dishes (Nunclon, Rochester, USA) in the presence of 100nM Red-Deox B 7,4 or vehicle control. After incubating the dishes for 7 days (AML) or 14 days (normal PBSCs) at 37°C in a humidified air atmosphere supplemented with 5% CO2, the numbers of AML colonies (CFU-L) containing at least 10 cells and normal PBSC colonies (BFU-E and CFU-GM) containing more than 100 cells were counted as previously described[3, 4]. To confirm the type of cells in the colonies, cells were picked when necessary and stained with May-Grϋnwald-Giemsa as described[3]. 3 BERNARD et al SELECT MICROTUBULE INHIBITORS DISRUPT LYSOSOMES In vivo xenograft studies Animal studies were carried out according to the regulations of the Canadian Council on Animal Care and with the approval of the Princess Margaret Cancer Centre ethics review board. Briefly, SCID mice (Ontario Cancer Institute, Toronto, ON, Canada) were injected subcutaneously with 5x105 OCI-AML-2 cells. Six days post-challenge, when tumors were palpable, mice were treated 5 days per week with twice-daily intraperitoneal injections of Red-Deox B 7,4 (-) (75mg/kg) or vehicle control (0.9% NaCl, 10% DMSO and 10% Cremophor EL). Mice were sacrificed 18 days post-tumor challenge. Cell cycle analysis Cells were treated with microtubule inhibitors for the specified times, harvested, washed once in cold PBS and fixed overnight in cold 70% ethanol at -20°C. Cell were then pelleted, washed once with PBS and treated with 100ng/ml DNase-free RNase A (Invitrogen, Carlsbad, CA) at 37°C for 30 minutes. A 5μg/ml Propidium Iodine (PI) solution was added prior to measuring DNA content on a FACScanto II (Becton Dickinson, FL). Results were analyzed with FlowJo (TreeStar, Ashland, OR). Mitotic block reversibility assay Mitotic block reversibility was assessed exactly as previously described[5]. Briefly, U937 cells were seeded in 75cm2 flasks at 105 cells/ml and allowed to resume log-phase growth. Concentrations of microtubule inhibitors ranging from 0.25X to 4X their respective IC50 values for U937 cells were added for 12 hours prior to washing the cells twice with pre-warmed PBS 4 BERNARD et al SELECT MICROTUBULE INHIBITORS DISRUPT LYSOSOMES and replenishing with warm, CO2-equilibrated drug-free media. Cells were allowed to recover for 5 days, with one 80% media replenishment on day 2. Cells were sampled for cell cycle analysis at the end of the 12 hour drug treatment period as well as 10 hours post-PBS. Lysosome enrichment The lysosome enrichment method was adapted from Lardeux et al[6]. Briefly, 108 cells per condition were harvested, washed once in cold PBS and resuspended in cold 0.25M sucrose buffer containing 10mM TRIS pH 7.4, 10mM KH2PO4, 0.5mM EDTA and protease inhibitors (cOmplete Mini tablets; Roche Applied Science, IN). Protease inhibitors were omitted when isolating lysosomes to assess cathepsin B release. All subsequent steps were carried out at 4°C and samples kept on ice. Cell homogenates were obtained using a teflon-glass homogenizer (Glas-Col, Terre Haute, IN) and centrifuged at 3000rpm for 10mins. Supernatants were next centrifuged at 5000 rpm for an additional 10 minutes. Lysosomes were purified from the resulting supernatants by centrifugation at 10000rpm for 20 minutes and resuspended in sucrose buffer supplemented with TRIS. Protein concentrations were determined using the Bradford reagent (Bio-Rad, Hercules, CA). V-ATPase gene expression analysis The expression of V-ATPase genes in the dataset GSE9476 was analyzed. This dataset contains gene expression data from 26 primary AML samples and 8 normal CD34+ samples[7]. Mean fold-change for the AML vs. normal CD34+ bone marrow sample comparison was calculated for all V-ATPase components present on the array (Affymetrix HG U133A GeneChip), and statistical significance was assessed by ANOVA. Lists of significantly differentially expressed 5 BERNARD et al SELECT MICROTUBULE INHIBITORS DISRUPT LYSOSOMES genes, including median fold-change and significance values for each gene, were thus derived, and visualized using a waterfall plot. REFERENCES 1. Hamilton TC, Young RC, Ozols RF (1984) Experimental model systems of ovarian cancer: applications to the design and evaluation of new treatment approaches. Semin Oncol 11:285–298. 2. Loganzo F, Hari M, Annable T, et al. (2004) Cells resistant to HTI-286 do not overexpress Pglycoprotein but have reduced drug accumulation and a point mutation in alpha-tubulin. Mol Cancer Ther 3:1319–1327. 3. Buick RN, Till JE, McCulloch EA (1977) Colony assay for proliferative blast cells circulating in myeloblastic leukaemia. Lancet 1:862–863. 4. Fauser AA, Messner HA (1979) Identification of megakaryocytes, macrophages, and eosinophils in colonies of human bone marrow containing neurtophilic granulocytes and erythroblasts. Blood 53:1023–1027. 5. Towle MJ, Salvato KA, Wels BF, et al. (2011) Eribulin induces irreversible mitotic blockade: implications of cell-based pharmacodynamics for in vivo efficacy under intermittent dosing conditions. Cancer Res 71:496–505. doi: 10.1158/0008-5472.CAN-10-1874 6. Lardeux B, Gouhot B, Forestier M (1983) Improved recovery of rat liver fractions enriched in lysosomes by specific alteration of the sedimentation properties of mitochondria. Anal Biochem 131:160–165. 7. Stirewalt DL, Meshinchi S, Kopecky KJ, et al. (2008) Identification of genes with abnormal expression changes in acute myeloid leukemia. Genes Chromosomes Cancer 47:8–20. doi: 10.1002/gcc.20500 6 BERNARD et al SELECT MICROTUBULE INHIBITORS DISRUPT LYSOSOMES FigS1. Deox B 7,4 promotes apoptosis in various AML cell lines in vitro. Cell death was determined at 72hrs using AnnexinV and PI staining following treatment of various AML cell lines with increasing concentrations of Deox B 7,4. 7 BERNARD et al SELECT MICROTUBULE INHIBITORS DISRUPT LYSOSOMES FigS2. Low oxygen levels increases HIF1 expression in TEX cells TEX cells were cultured for 24 hours with 21% or 0.2% oxygen. After incubation, cells were harvested and HIF1 mRNA expression as measured by Q-RTPCR. 8 BERNARD et al SELECT MICROTUBULE INHIBITORS DISRUPT LYSOSOMES Supplementary FigS3A. Red-Deox B 7,4 (-) toxicology in vivo (biochemistry). Serum levels of liver enzymes (AST, ALP), creatine/creatine kinase and bilirubin in SCID mice after 12 days of Red-Deox B 7,4 (-) administration (75mg/kg intraperitoneally, twice daily, 5 days per week). Lines represent median. 9 BERNARD et al SELECT MICROTUBULE INHIBITORS DISRUPT LYSOSOMES Supplementary FigS3B. Red-Deox B 7,4 (-) toxicology in vivo (histology). SCID mice were treated for 12 days with Red-Deox B 7,4 (-) (75mg/kg intraperitoneally, twice daily, 5 days per week) or vehicle control. After treatment, mice were sacrificed, organs harvested and stained with hematoxylin and eosin. 10 V-ATPase activity (acridine orange fluorescence) BERNARD et al SELECT MICROTUBULE INHIBITORS DISRUPT LYSOSOMES Control 4hrs Deox B 7,4 0 8hrs Deox B 7,4 -50 -100 -150 -200 0 60 120 180 Time (secs) Supplementary FigS4. Deox B 7,4 leads to increased V-ATPase activity. V-ATPase activity was measured using an acridine orange quenching assay on lysosomes purified from TEX cells treated for 4hrs or 8hrs with 2.5X the IC50 value for Deox B 7,4 or for 8hrs with DMSO (refer to Table S2 for IC50 value). Fluorescence was measured every 20secs for 3mins using excitation and emission wavelengths of 490nm and 540nm, respectively. Results shown have been baseline-corrected and are representative of 2 experiments. 11 BERNARD et al SELECT MICROTUBULE INHIBITORS DISRUPT LYSOSOMES % Lysosomal integrity Deox B 7,4 R-Deox B 7,4 (-) Nocodazole Colchicine Vinblastine Vincristine Paclitaxel Bafilomycin A1 100 90 80 60 40 20 0 2 4 8 16 24 Time (hrs) Supplementary FigS5. R-Deox B 7,4 (-) promotes lysosomal disruption in OCI-AML-2 cells OCI-AML-2 cells were treated with various microtubule inhibitors for the specified times using concentrations equivalent to 2.5X their respective IC50 values for this cell line (refer to Table S2 for IC50 values). Lysosome integrity was measured using acridine orange staining and flow cytometry. Bafilomycin A1 was used as a positive control. Only viable cells, based on forward and side scatter properties, were included in the analysis. 12 SELECT MICROTUBULE INHIBITORS DISRUPT LYSOSOMES 10000 *** 7500 5000 2500 0 R -D eo C on tr x ol B 7, N oc 4 ( ) od az ol C ol e ch ic Vi nb ine B af las tin ilo e m yc in A Tr 1 ito nX Cathepsin B release (relative fluorescence units) BERNARD et al Supplementary FigS6. Deox B 7,4 and Red-Deox B 7,4 (-) do not affect lysosome integrity directly. Lysosomes were isolated from TEX cells and treated with various microtubule inhibitors as well as bafilomycin A1 (40μg/sample) for 2hrs before assessing cathepsin B release. Triton-X detergent was used as a positive control. Drug concentrations used were equivalent to 10X their respective IC50 values for this cell line (refer to Table S2 for IC50 values). Results are shown as mean 95% CI and are representative of 2 experiments (***p<0.0001 compared to control cells). 13 BERNARD et al SELECT MICROTUBULE INHIBITORS DISRUPT LYSOSOMES Supplementary Table S1. List of IC50 values for the various drugs used in this study Drug TEX [nM] OCI-AML-2 [nM] U937 [nM]* 325.67 ± 11.33 208.33 ± 15.86 554.13 ± 48.44 Red-Deox B 7,4 (-) Nocodazole Colchicine Vinblastine 41.21 ± 2.14 45.17 ± 4.53 9.15 ± 0.45 2.02 ± 0.32 26.60 ± 1.4 46.44 ± 3.75 9.72 ± 0.39 1.04 ± 0.11 328.89 ± 20.88 n/a 5.47 ± 0.25 0.68 ± 0.032 Vincristine Paclitaxel 4.13 ± 0.25 2.64 ± 0.20 3.54 ± 1.39 0.056 ± 0.06 n/a n/a Bafilomycin A1 23.05 ± 1.87 3.71 ± 0.39 n/a Deox B 7,4 IC50 values represent the mean 95% CI obtained from a minimum of 3 independent experiments. *IC50 values for U937 cells represent the mean 95% CI obtained from triplicate values of one experiment (n/a: not assessed). 14 BERNARD et al SELECT MICROTUBULE INHIBITORS DISRUPT LYSOSOMES Supplementary Table S2. Patient characteristics of primary AML samples Patient Gender Age 1 2 3 4 5 M F F M M 72 74 67 80 62 Disease status Cytogenetics Molecular AML with MDS related changes AML NOS- AML M5 AML NOS- AML M1 AML NOS- AML M5 AML NOS- AML M5 > 4 abnormalities including -5, -7, -17 XX XX Trisomy 8, trisomy 15 XY Not tested Not tested FLT3-ve, NPM-ve Not tested FLT3-ve, NPM-ve 15 BERNARD et al SELECT MICROTUBULE INHIBITORS DISRUPT LYSOSOMES Supplementary Table S3. List of S. cerevisiae screen hits obtained with Deox B 7,4 in glycolytic growth conditions (YPD medium) and respiratory growth conditions (YPGE medium) Gene 3.353 Systematic name YPL254W Glycolytic growth conditions (YPD medium) Standard name Brief description HFI1 Histone H2A Functional Interactor 3.226 3.012 YPR135W YOR153W CTF4 PDR5 Chromosome Transmission Fidelity Pleiotropic Drug Resistance 2.942 YDL185W VMA1 Vacuolar Membrane Atpase 2.881 YPR124W CTR1 Copper TRansport 2.842 2.785 YLR208W YLR396C SEC13 VPS33 SECretory Vacuolar Protein Sorting 2.661 2.609 2.511 2.135 YDL152W YKL118W YLR103C YGR180C CDC45 RNR4 Cell Division Cycle RiboNucleotide Reductase 2.046 YGR252W GCN5 General Control Nonderepressible 2.043 YOR281C PLP2 Phosducin-Like Protein 2.018 YKL119C VPH2 Vacuolar pH 1.931 1.922 1.842 YHR062C YER155C YBR118W RPP1 BEM2 TEF2 Ribonuclease P Protein Bud EMergence Translation Elongation Factor Log2 ratio Adaptor protein required for structural integrity of the SAGA complex Chromatin-associated protein Plasma membrane ATP-binding cassette (ABC) transporter Subunit A of the V1 peripheral membrane domain of VATPase High-affinity copper transporter of the plasma membrane Structural component of 3 distinct complexes ATP-binding protein that is a subunit of the HOPS and CORVET complexes Dubious open reading frame Dubious open reading frame DNA replication initiation factor Ribonucleotide-diphosphate reductase (RNR) small subunit Catalytic subunit of ADA and SAGA histone acetyltransferase complexes Protein that interacts with the CCT complex to stimulate actin folding Integral membrane protein required for V-ATPase function Subunit of both RNase MRP and nuclear RNase P Rho GTPase activating protein (RhoGAP) Translational elongation factor EF-1 alpha 16 BERNARD et al SELECT MICROTUBULE INHIBITORS DISRUPT LYSOSOMES 1.815 YGL005C COG7 Conserved Oligomeric Golgi complex RecQ Mediated genome Instability 1.814 YPL024W RMI1 1.811 1.786 1.703 1.699 YDR288W YJL184W YOR224C YOR098C NSE3 GON7 RPB8 NUP1 Non SMC Element 1.674 YHR083W SAM35 Sorting and Assembly Machinery 1.656 YNL139C THO2 1.602 1.591 1.573 1.565 YIL021W YJL009W YHR101C YBL042C RPB3 suppressor of the Transcriptional defect of Hpr1 by Overexpression RNA Polymerase B BIG1 FUI1 Bad In Glucose 5-FlUorourIdine resistance 1.534 YCR084C TUP1 1.490 YOR035C SHE4 deoxyThymidine monophosphate UPtake Swi5p-dependent HO Expression 1.452 YKL037W AIM26 1.446 YLR029C RPL15A 1.439 1.426 1.420 1.395 YOR340C YML032C YNL225C YLR240W RPA43 RAD52 CNM67 VPS34 RNA Polymerase B NUclear Pore Altered Inheritance rate of Mitochondria Ribosomal Protein of the Large subunit RNA Polymerase A RADiation sensitive Chaotic Nuclear Migration Vacuolar Protein Sorting Component of the conserved oligomeric Golgi complex Subunit of the RecQ (Sgs1p) - Topo III (Top3p) complex Component of the SMC5-SMC6 complex Component of the EKC/KEOPS protein complex RNA polymerase subunit ABC14.5 FG-nucleoporin component of central core of the nuclear pore complex Component of the sorting and assembly machinery (SAM) complex Subunit of the THO complex RNA polymerase II third largest subunit B44 Dubious open reading frame Integral membrane protein of the endoplasmic reticulum High affinity uridine permease, localizes to the plasma membrane General repressor of transcription Protein containing a UCS (UNC-45/CRO1/SHE4) domain Putative protein of unknown function Ribosomal 60S subunit protein L15A RNA polymerase I subunit A43 Protein that stimulates strand exchange Component of the spindle pole body outer plaque Phosphatidylinositol (PI) 3-kinase that synthesizes PI-3phosphate 17 BERNARD et al SELECT MICROTUBULE INHIBITORS DISRUPT LYSOSOMES 1.384 YHR024C MAS2 Mitochondrial ASsembly 1.380 YDL139C SCM3 1.366 1.363 YDL150W YMR309C RPC53 NIP1 Suppressor of Chromosome Missegregation RNA Polymerase C Nuclear ImPort 1.350 1.339 1.328 YOL071W YLR336C YOR136W EMI5 SGD1 IDH2 Early Meiotic Induction Suppressor of Glycerol Defect Isocitrate DeHydrogenase 1.308 YAL010C MDM10 1.304 YDR359C EAF1 Mitochondrial Distribution and Morphology Esa1p-Associated Factor 1.295 1.286 1.257 1.255 1.247 1.237 YPL142C YDR317W YER058W YNL199C YMR168C YLL037W HIM1 PET117 GCR2 CEP3 High Induced Mutagenesis PETite colonies GlyColysis Regulation CEntromere Protein 1.225 YJL209W CBP1 Cytochrome B mRNA Processing 1.205 1.194 1.184 YBL041W YLR010C YGR020C PRE7 TEN1 VMA7 PRoteinase yscE TElomeric pathways with STn1 Vacuolar Membrane Atpase 1.170 1.168 YDR381W YDR331W YRA1 GPI8 Yeast RNA Annealing protein GlycosylPhosphatidylInositol anchor biosynthesis Larger subunit of the mitochondrial processing protease (MPP) Nonhistone component of centromeric chromatin RNA polymerase III subunit C53 eIF3c subunit of the eukaryotic translation initiation factor 3 (eIF3) Subunit of succinate dehydrogenase Essential nuclear protein Subunit of mitochondrial NAD(+)-dependent isocitrate dehydrogenase Subunit of both the ERMES complex and the SAM complex Component of the NuA4 histone acetyltransferase complex Dubious open reading frame Protein of unknown function involved in DNA repair Protein required for assembly of cytochrome c oxidase Transcriptional activator of genes involved in glycolysis Essential kinetochore protein Dubious open reading frame unlikely to encode a functional protein Mitochondrial protein, regulator of COB mRNA stability and translation Beta 6 subunit of the 20S proteasome Protein that regulates telomeric length Subunit F of the V1 peripheral membrane domain of VATPase Nuclear polyadenylated RNA-binding protein ER membrane glycoprotein subunit of the GPI transamidase complex 18 BERNARD et al SELECT MICROTUBULE INHIBITORS DISRUPT LYSOSOMES 1.163 YOR073W SGO1 1.162 YNL131W TOM22 1.158 YNL256W FOL1 ShuGOshin (Japanese for "guardian spirit") Translocase of the Outer Mitochondrial membrane FOLic acid synthesis 1.151 1.149 YLR293C YAL016W GSP1 TPD3 Genetic Suppressor of Prp20-1 tRNA Processing Deficient 1.140 1.137 1.126 1.109 1.097 1.066 1.063 YFR050C YBR289W YDR280W YDL143W YMR205C YLR274W YPR134W PRE4 SNF5 RRP45 CCT4 PFK2 MCM5 MSS18 PRoteinase yscE Sucrose NonFermenting Ribosomal RNA Processing Chaperonin Containing TCP-1 PhosphoFructoKinase MiniChromosome Maintenance Mitochondrial Splicing System 1.058 1.055 YOR302W YOL133W HRT1 1.048 1.047 1.047 1.046 1.040 1.029 YFL037W YCL017C YDR045C YJL001W YDL092W YPR132W TUB2 NFS1 RPC11 PRE3 SRP14 RPS23B 1.028 1.024 YDR232W YOR209C HEM1 NPT1 High level expression Reduces Ty3 transposition TUBulin NiFS-like RNA Polymerase C PRoteinase yscE Signal Recognition Particle Ribosomal Protein of the Small subunit HEMe biosynthesis Nicotinate PhosphoribosylTransferase 1.023 YLR076C Component of the spindle checkpoint Component of the TOM (Translocase of Outer Membrane) complex Multifunctional enzyme of the folic acid biosynthesis pathway Ran GTPase Regulatory subunit A of the heterotrimeric PP2A complex Beta 7 subunit of the 20S proteasome Subunit of the SWI/SNF chromatin remodeling complex Exosome non-catalytic core component Subunit of the cytosolic chaperonin Cct ring complex Beta subunit of heterooctameric phosphofructokinase Component of the Mcm2-7 hexameric helicase complex Nuclear encoded protein needed for splicing of mitochondrial intron CPA1 uORF RING-H2 domain core subunit of multiple ubiquitin ligase complexes Beta-tubulin Cysteine desulfurase RNA polymerase III subunit C11 Beta 1 subunit of the 20S proteasome Signal recognition particle (SRP) subunit Ribosomal protein 28 (rp28) of the small (40S) ribosomal subunit 5-aminolevulinate synthase Nicotinate phosphoribosyltransferase Dubious open reading frame 19 BERNARD et al SELECT MICROTUBULE INHIBITORS DISRUPT LYSOSOMES 1.013 YJL026W RNR2 RiboNucleotide Reductase Ribonucleotide-diphosphate reductase (RNR), small subunit Mitochondrial oxidoreductase 1.013 YKL195W MIA40 1.006 YKL059C MPE1 1.005 YOR358W HAP5 Log2 ratio 4.102 2.998 Systematic name YCR009C YKL119C Gene Mitochondrial intermembrane space Import and Assembly Mutant PCF11 Extragenic suppressor Essential conserved subunit of CPF cleavage and polyadenylation factor Heme Activator Protein Subunit of the Hap2p/3p/4p/5p CCAAT-binding complex Respiratory growth conditions (YPGE medium) Standard name Brief description RVS161 VPH2 Reduced Viability on Starvation Vacuolar pH 2.688 YNR036C MRPS12 2.530 2.528 2.404 2.299 2.181 YPL118W YDR477W YLR270W YGL240W YNL243W MRP51 SNF1 DCS1 DOC1 SLA2 Mitochondrial Ribosomal Protein, Small subunit Mitochondrial Ribosomal Protein Sucrose NonFermenting DeCapping Scavenger Destruction Of Cyclin B Synthetic Lethal with ABP1 2.106 YGL115W SNF4 1.957 1.875 1.818 1.729 1.720 1.656 YOR333C YPL031C YJL140W YDL090C YJR105W YKL080W PHO85 RPB4 RAM1 ADO1 VMA5 Amphiphysin-like lipid raft protein Integral membrane protein required for V-ATPase function Mitochondrial ribosomal protein of the small subunit Mitochondrial ribosomal protein of the small subunit AMP-activated serine/threonine protein kinase Non-essential hydrolase involved in mRNA decapping Processivity factor Adaptor protein that links actin to clathrin and endocytosis Sucrose NonFermenting Activating gamma subunit of the AMP-activated Snf1p kinase complex Dubious open reading frame PHOsphate metabolism Cyclin-dependent kinase RNA Polymerase B RNA polymerase II subunit B32 RAS protein and A-factor Maturation Beta subunit of the CAAX farnesyltransferase (FTase) ADenOsine kinase Adenosine kinase Vacuolar Membrane Atpase Subunit C of the V1 peripheral membrane domain of VATPase 20 BERNARD et al SELECT MICROTUBULE INHIBITORS DISRUPT LYSOSOMES 1.654 YIL134W FLX1 FLavin eXchange 1.612 YMR188C MRPS17 1.607 1.588 1.551 1.547 YJR104C YGL148W YDR008C YGL244W SOD1 ARO2 Mitochondrial Ribosomal Protein, Small subunit SuperOxide Dismutase AROmatic amino acid requiring RTF1 Restores TBP Function 1.546 YER070W RNR1 RiboNucleotide Reductase 1.520 YBL099W ATP1 ATP synthase 1.502 1.476 1.398 YNR025C YMR077C YGR020C VPS20 VMA7 Vacuolar Protein Sorting Vacuolar Membrane Atpase 1.345 1.307 1.304 YBR171W YDL006W YOR209C SEC66 PTC1 NPT1 1.280 YOR332W VMA4 SECretory Phosphatase type Two C Nicotinate PhosphoribosylTransferase Vacuolar Membrane Atpase 1.254 YPR173C VPS4 Vacuolar Protein Sorting 1.240 YDR359C EAF1 Esa1p-Associated Factor 1.234 1.227 1.213 YLR038C YBR026C YEL027W COX12 ETR1 VMA3 Cytochrome c OXidase 2-Enoyl Thioester Reductase Vacuolar Membrane Atpase Protein required for transport of flavin adenine dinucleotide (FAD) Mitochondrial ribosomal protein of the small subunit Cytosolic copper-zinc superoxide dismutase Bifunctional chorismate synthase and flavin reductase Dubious open reading frame Subunit of RNAPII-associated chromatin remodeling Paf1 complex Major isoform of large subunit of ribonucleotidediphosphate reductase Alpha subunit of the F1 sector of mitochondrial F1F0 ATP synthase Dubious open reading frame Myristoylated subunit of ESCRTIII Subunit F of the V1 peripheral membrane domain of VATPase Non-essential subunit of Sec63 complex Type 2C protein phosphatase (PP2C) Nicotinate phosphoribosyltransferase Subunit E of the V1 domain of the vacuolar H+-ATPase (V-ATPase) AAA-ATPase involved in multivesicular body (MVB) protein sorting Component of the NuA4 histone acetyltransferase complex Subunit VIb of cytochrome c oxidase 2-enoyl thioester reductase Proteolipid subunit c of the V0 domain of vacuolar H(+)-ATPase 21 BERNARD et al SELECT MICROTUBULE INHIBITORS DISRUPT LYSOSOMES 1.200 1.200 1.189 YDR245W YGL206C YER122C MNN10 CHC1 GLO3 MaNNosyltransferase Clathrin Heavy Chain GLyOxalase 1.185 YNL112W DBP2 Dead Box Protein 1.158 1.151 YDR264C YMR123W AKR1 PKR1 1.132 YOR196C LIP5 AnKyrin Repeat-containing protein Pichia farinosa Killer toxin Resistance LIPoic acid 1.128 YPL078C ATP4 ATP synthase 1.107 1.095 1.053 1.041 1.024 1.024 YLR260W YDL206W YHL025W YIL060W YGL168W YGR105W LCB5 Long-Chain Base SNF6 Sucrose NonFermenting HUR1 VMA21 HydroxyUrea Resistance Vacuolar Membrane Atpase 1.011 YOR014W RTS1 Rox Three Suppressor 1.007 YMR058W FET3 FErrous Transport Subunit of a Golgi mannosyltransferase complex Clathrin heavy chain ADP-ribosylation factor GTPase activating protein (ARF GAP) ATP-dependent RNA helicase of the DEAD-box protein family Palmitoyl transferase involved in protein palmitoylation V-ATPase assembly factor Protein involved in biosynthesis of the coenzyme lipoic acid Subunit b of the stator stalk of mitochondrial F1F0 ATP synthase Minor sphingoid long-chain base kinase Putative protein of unknown function Subunit of the SWI/SNF chromatin remodeling complex Putative protein of unknown function Protein of unknown function Integral membrane protein required for V-ATPase function B-type regulatory subunit of protein phosphatase 2A (PP2A) Ferro-O2-oxidoreductase Haplo-insufficiency profiling (HIP) of Deox B 7,4 in S. cerevisiae was performed in either glycolytic growth conditions (YPD medium) or respiratory growth conditions (YPGE medium). The fitness defects were calculated for each deletion strain as log2 ratios (mean signal intensity of control/mean signal intensity of drug). Deletion strains with log2 ratio values of more than 1 were considered hits. 22 BERNARD et al SELECT MICROTUBULE INHIBITORS DISRUPT LYSOSOMES Supplementary Table S4. Microtubule inhibition by Deox B 7,4 and R-Deox B 7,4 (-) is functionally relevant and overcomes drug efflux pump-related resistance Drug KB-3-1 [nM] Deox B 7,4 R-Deox B 7,4 Nocodazole Colchicine 344.46 ± 26.34 89.30 ± 23.32 40.35 ± 3.92 6.14 ± 0.12 KB-4.0-HTI [nM] 655.93 ± 31.46 149.72 ± 29.83 99.79 ± 10.85 17.57 ± 1.06 Fold resistance (KB4.0/KB3.1) 1.90 1.68 2.47 2.86 Vincristine n/a n/a n/a Doxorubicin n/a n/a n/a A2780 [nM] 300.83 ± 70.23 39.50 ± 0.71 n/a 5.47 ± 0.12 A2780ADR [nM] 576.17 ± 101.87 146.30 ± 42.19 n/a 324.26 ± 39.14 Fold resistance (A2780ADR/A2780) 1.92 3.70 n/a 59.28 0.77 ± 0.09 15.59 ± 8.83 845.74 ± 205.67 729.57 ± 148.45 1098.36 46.80 Alpha-tubulin mutated KB-4.0-HTI epidermoid carcinoma cells and their KB-3-1 wild-type controls, as well as A2780ADR ovarian carcinoma cells overexpressing P-glycoprotein and their A2780 wild-type controls, were treated with increasing concentrations of the specified microtubule inhibitors for 72hrs. Cell viability was assessed using MTS and data represent IC 50 values ± standard deviation calculated from 3 individual experiments (n/a: not assessed).sss 23