IGES-2015
advertisement

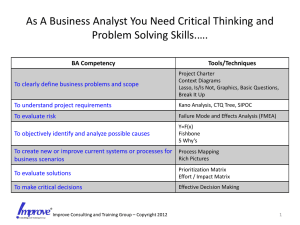
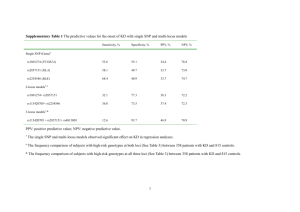
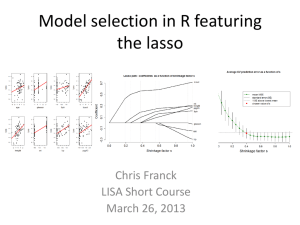
LASSO-Based Approaches for Joint Tests of Genetic Main Effects and Gene–Environment Interactions Jie Zheng1, Dabeeru C. Rao2, Gang Shi3* 1Clinical Research Center, The First Affiliated Hospital of X’an Jiaotong University, 227 West Yanta Road, Xi’an, Shaanxi, 710061, China, jie_stat33@yahoo.com 2Division of Biostatistics, Washington University School of Medicine, 660 South Euclid Avenue, Campus Box 8067, Saint Louis, Missouri, 63110, USA, rao@wubios.wustl.edu 3State Key Laboratory of Integrated Services Networks, Xidian University, 2 South Taibai Road, Xi’an, Shaanxi, 710071, China, gshi@xidian.edu.cn *Corresponding author Abstract The least absolute shrinkage and selection operator (LASSO) regression, the regression coefficients of which are under an L1 norm constraint, has the so-called shrinkage property and yields a parsimonious model. It is an appealing approach for variable selection, especially for high-dimensional problems, and has attracted considerable interest in genetic studies. The LASSO regression and its variations have been applied to screening common and rare genetic variants (Shi et al., 2011; Zhou et al., 2010), testing single-nucleotide polymorphisms (SNPs) (Ayers et al., 2011) and haplotypes (Biswas and Lin, 2012; Lin et al., 2012), detecting SNPSNP interactions (D'Angelo et al., 2009; Wu et al, 2010) and haplotypehaplotype interactions (Li et al., 2010), jointly examining genetic main and dominant effects (Sabourin et al., 2015), estimating gene–gene and gene–environment interactions simultaneously (Tanck et al., 2006), analyzing main effects of SNPs and their epistatic interactions jointly (Yang et al., 2010). In this work, we investigated application of the LASSO regression to the joint testing of genetic main effects and gene–environment interactions considering multiple SNPs simultaneously. Testing genetic main effects together with interaction effects is known to be more powerful than either of the marginal tests, if an interaction exists. A recent large-scale, genome-wide association study of blood pressure to assess the pervasiveness of gene–age interactions employed the two degrees-of-freedom joint test and identified twenty significant loci, of which nine demonstrated nominal evidence of interactions with age and five would have been missed by a main-effects-only analysis. We generalized the single-marker two degrees-of-freedom joint test to a multiple-marker version using the LASSO regression. Specifically, we considered two LASSO-based approaches. One analyzed the genetic main effects of multiple SNPs and their interactions with an environmental factor. SNPs with a nonzero regression coefficient for either a main effect or interaction were considered significant. The other employed the group LASSO regression, which shrinks each SNP with its interaction in a group-wise manner. SNPs in the final model were deemed significant (for both main effects and interactions). The two degrees-of-freedom linear regression that tests the main effects and interactions jointly was used as the benchmark test for comparisons. The statistical power for testing SNPs with various main and interaction effect sizes was evaluated, and type I errors of the three methods were examined. Bayesian information criterion (BIC), Mallow's Cp, and Stine's Sp were compared with respect to their ability to select constraint parameters of the two LASSO approaches. Based on simulation studies, we showed that BIC and Mallow's Cp tend to generate over-fitted models resulting in large type I errors for both the LASSO methods. Choosing constraint parameters based on Stine's Sp demonstrated acceptable empirical type I errors. Since Stine's Sp balances the model fitness and number of parameters and does not control type I errors directly, the two LASSO methods showed varying empirical false positive rates for different sample sizes. Group LASSO displayed lower type I errors and statistical power than did the two degrees-of-freedom regression test when sample sizes were relatively small and higher type I errors and statistical power when sample sizes were larger. The first LASSO method showed higher type I errors and statistical power than did the group LASSO and had a similar pattern of type I errors for different sample sizes. For computational efficiency, the first approach was much faster than the second when using the least-angle regression solver. Reference Ayers KL, Cordell HJ. SNP selection in genome-wide and candidate gene studies via penalized logistic regression. Genet Epidemiol. 2010;34(8):879-91. doi: 10.1002/gepi.20543. Biswas S, Lin S. Logistic Bayesian LASSO for identifying association with rare haplotypes and application to age-related macular degeneration. Biometrics. 2012;68(2):587-97. doi: 10.1111/j.1541-0420.2011.01680.x. Li M, Romero R, Fu WJ, Cui Y. Mapping haplotype-haplotype interactions with adaptive LASSO. BMC Genet. 2010;11:79. doi: 10.1186/1471-2156-11-79. Lin WY, Yi N, Zhi D, Zhang K, Gao G, Tiwari HK, Liu N. Haplotype-based methods for detecting uncommon causal variants with common SNPs. Genet Epidemiol. 2012;36(6):572-82. doi: 10.1002/gepi.21650. Sabourin J, Nobel AB, Valdar W. Fine-mapping additive and dominant SNP effects using group-LASSO and fractional resample model averaging. Genet Epidemiol. 2015;39(2):77-88. doi: 10.1002/gepi.21869. Shi G, Boerwinkle E, Morrison AC, Gu CC, Chakravarti A, Rao DC. Mining gold dust under the genome wide significance level: a two-stage approach to analysis of GWAS. Genet Epidemiol. 2011; 35(2):111-118. doi: 10.1002/gepi.20556. Tanck MW, Jukema JW, Zwinderman AH. Simultaneous estimation of gene-gene and gene-environment interactions for numerous loci using double penalized log-likelihood. Genet Epidemiol. 2006;30(8):645-51. Wu J, Devlin B, Ringquist S, Trucco M, Roeder K. Screen and clean: a tool for identifying interactions in genome-wide association studies. Genet Epidemiol. 2010;34(3):275-85. doi: 10.1002/gepi.20459. Yang C, Wan X, Yang Q, Xue H, Yu W. Identifying main effects and epistatic interactions from large-scale SNP data via adaptive group Lasso. BMC Bioinformatics. 2010;11 Suppl 1:S18. doi: 10.1186/1471-2105-11-S1-S18. Zhou H, Sehl ME, Sinsheimer JS, Lange K. Association screening of common and rare genetic variants by penalized regression. Bioinformatics. 2010;26(19):2375-82. doi: 10.1093/bioinformatics/btq448.