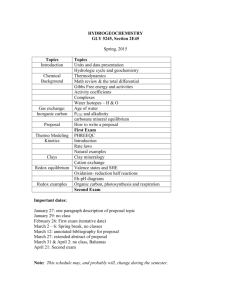
Table S2-1. Rate equations for Model ii
advertisement

Supporting Information Kinetic Modeling of Pt/C Catalyzed Aqueous Phase Glycerol Conversion with In Situ Formed Hydrogen Xin Jin1, Prem S. Thapa2, Bala Subramaniam1, Raghunath V. Chaudhari1* 1 Center for Environmentally Beneficial Catalysis, Department of Chemical and Petroleum Engineering, University of Kansas, 1501 Wakarusa Drive, Lawrence, Kansas 66047, USA 2 Microscopy and Analytical Imaging Laboratory, Haworth Hall, 1200 Sunnyside Ave, University of Kansas, Lawrence, Kansas 66045, USA *To whom the correspondence should be addressed. E-mail: rvc1948@ku.edu KEYWORDS Kinetic modeling, Polyol, Tandem catalysis, Dehydrogenation, Hydrogenolysis 1 (a) (b) Figure S1. (a) Temporal glycerol concentration profile at different temperatures, and (b) apparent first order dependence (PN2: 1.4 MPa, glycerol: 1.1 kmol/m3, solvent: H2O, NaOH/glycerol molar ratio: 1.1, Pt/C catalyst charge: 6.7 kg/m3) Figure S2. Apparent activation energy for glycerol conversion on Pt/C catalyst 2 (a) (b) (c) Figure S3. Arrhenius and van’t Hoff plots of rate and adsorption equilibrium constants on Pt/C catalyst 3 Figure S4. Concentration-time profiles of glycerol conversion on Pt/C catalyst at 130 oC with different initial glycerol and NaOH concentration 4 Figure S5. Concentration-time profiles of glycerol conversion on Pt/C catalyst at 160 oC with different initial NaOH concentration 5 1. Derivation of Models ii~vi Table S1-1. Derivation of surface reactions in Model ii No. Surface reaction Rate equation r1 r1 r2 r2 r3 r3 1 K k s1 K gly C gly K OH COH C gly K EG CEG K OH COH 2 gly 1 K k s 2 K gly C gly K OH COH C gly K EG C EG K OH COH 2 gly k s 3 K1, 2PDO C1, 2PDO KOH COH 1 K 1, 2 PDO r4 r4 r5 r5 r6 r6 6 2 k s 4 K gly C gly K OH C OH 1 K 1 K C1, 2PDO C gly K EG C EG K OH C OH 2 gly k s 5 K gly C gly K OH C OH C gly K EG C EG K OH C OH 2 gly 1 K k s 6 K EG C EG K OH C OH C gly K EG C EG K OH C OH 2 gly Table S1-2. Derivation of surface reactions in Model iii No. Surface reaction Rate equation r1 r1 r2 r2 r3 r3 r4 r4 r5 r5 r6 r6 7 k s1 K gly C gly COH 1 K gly C gly K OH COH 2 k s 2 K gly C gly COH 1 K C gly K OH COH 2 gly k s 3 K1, 2 PDO C1, 2 PDO COH 1 K C1, 2 PDO 2 1, 2 PDO k s 4 K gly C gly COH 1 K C gly K OH COH 2 gly k s 5 K gly C gly COH 1 K C gly K OH COH 2 gly k s 6 K gly C gly COH 1 K gly C gly K OH COH 2 Table S1-3. Derivation of surface reactions in Model iv No. Surface reaction Rate equation r1 r1 r2 r2 r3 r3 1 K 1 K k s1 K gly Cgly COH Cgly K EG CEG 1 KOH COH 2 gly k s 2 K gly C gly COH C gly K EG C EG 1 K OH COH 2 gly k s 3 K1, 2PDO C1, 2PDO COH 1 K r4 r4 r5 r5 r6 r6 8 1 K 1 K 1 K C1, 2PDO 2 1, 2 PDO k s 4 K gly C gly COH C gly K EG C EG 1 K OH COH 2 gly k s 5 K gly C gly COH C gly K EG C EG 1 K OH C OH 2 gly k s 5 K EG C EG COH C gly K EG C EG 1 K OH COH 2 gly Table S1-4. Derivation of surface reactions in Model v No. Surface reaction Rate equation r1 r1 r2 r2 r3 r3 r4 r4 r5 r5 r6 r6 9 1 K k s1 K gly C gly K OH COH C gly K EG C EG 1 K OH COH 2 gly 1 K k s 2 K gly C gly K OH COH C gly K EG C EG 1 K OH COH 2 gly k s 3 K1, 2 PDO C1, 2 PDO K OH COH 1 K 1 K 1 K 1 K C1, 2 PDO 2 1, 2 PDO k s 4 K gly C gly K OH COH C gly K EG C EG 1 K OH COH 2 gly k s 5 K gly C gly K OH COH C gly K EG C EG 1 K OH COH 2 gly k s 5 K EG C EG K OH COH C gly K EG C EG 1 K OH COH 2 gly Table S1-5. Derivation of surface reactions in Model vi No. Surface reaction Rate equation r1 r1 r2 r2 r3 r3 1 K 1 K k s1 K gly C gly C OH C gly 1 K OH C OH 2 gly k s 2 K gly C gly COH C gly 1 K OH COH 2 gly k s 3 K1, 2PDO C1, 2PDO COH 1 K C1, 2PDO 2 1, 2 PDO r4 r4 r5 r5 r6 r6 10 1 K 1 K 1 K k s 4 K gly C gly COH C gly 1 K OH COH 2 gly k s 5 K gly C gly COH C gly 1 K OH COH 2 gly k s 5 K gly C gly COH C gly 1 K OH COH 2 gly 2. Data fitting for Models ii~vi Table S2-1. Rate equations for Model ii r1 r2 r3 1 K k s1 K gly C gly K OH COH C gly K EG CEG K OH COH 2 gly 1 K k s 2 K gly C gly K OH COH C gly K EG C EG K OH COH 2 gly k s 3 K1, 2PDO C1, 2PDO KOH COH 1 K r4 r5 r6 1 K 1 K 1 K C1, 2PDO 2 1, 2 PDO k s 4 K gly C gly K OH C OH C gly K EG C EG K OH C OH 2 gly k s 5 K gly C gly K OH C OH C gly K EG C EG K OH C OH 2 gly k s 6 K EG C EG K OH C OH C gly K EG C EG K OH C OH 2 gly Table S2-2. Parameter estimation for Model ii Model ii Constants ks1 ks2 ks3×10-1 ks4 ks5 ks6×103 Kgly×10-2 KOH-×10-2 K1,2-PDO KEG×10-3 130oC 145oC 160oC 44.0±3.17 20.0±1.66 3.76±0.89 3.68±0.43 <0 1.95±0.57 6.06 2.05 8.31±7.12 7.54 6.61±1.69 3.72±0.95 1.37±0.44 0.43±0.18 0.37±0.14 ~0 14.2 14.5 27.0±29.4 25.5±22.8 14.3±1.48 1.29 28.9±5.83 0.56±0.07 3.49±0.26 ~0 17.5 11.6 16.5 >>4000 11 Table S2-3. Rate equations for Model iii r1 r2 r3 r4 r5 r6 k s1 K gly C gly COH 1 K gly C gly K OH COH 2 k s 2 K gly C gly COH 1 K C gly K OH COH 2 gly k s 3 K1, 2 PDO C1, 2 PDO COH 1 K C1, 2 PDO 2 1, 2 PDO k s 4 K gly C gly COH 1 K C gly K OH COH 2 gly k s 5 K gly C gly COH 1 K C gly K OH COH 2 gly k s 6 K gly C gly COH 1 K gly C gly K OH COH 2 Table S2-4. Parameter estimation for Model iii Model iii Constants ks1 ks2×10-1 ks3×10-2 ks4×10-2 ks5×10-2 ks6×10-2 Kgly×10-2 KOH-×10-3 K1,2-PDO 130oC 145oC 160oC 1.33±0.10 6.05±0.53 6.47±0.96 6.75 9.77±1.67 3.42±1.61 3.80 1.00 0.38 1.98±0.16 10.8 24.8±3.37 11.7±1.5 24.4±1.68 >>600 3.75±0.22 ~0 0.20 10.2±0.42 5.32 241±16.9 23.0±2.77 144±11.9 2105±194 2.93±1.23 5.70±26.5 2.65 12 Table S2-5. Rate equations for Model iv r1 r2 r3 1 K k s1 K gly Cgly COH Cgly K EG CEG 1 KOH COH 2 gly 1 K k s 2 K gly C gly COH C gly K EG C EG 1 K OH COH 2 gly k s 3 K1, 2PDO C1, 2PDO COH 1 K r4 r5 r6 1 K 1 K 1 K C1, 2PDO 2 1, 2 PDO k s 4 K gly C gly COH C gly K EG C EG 1 K OH COH 2 gly k s 5 K gly C gly COH C gly K EG C EG 1 K OH COH 2 gly k s 5 K EG C EG COH C gly K EG C EG 1 K OH COH 2 gly Table S2-6. Parameter estimation for Model iv Model iv Constants ks1 ks2×10-1 ks3×10-2 ks4×10-2 ks5×10-2 ks6 Kgly×10-2 KOH-×10-3 K1,2-PDO KEG×10-2 130oC 145oC 160oC 1.28±0.10 5.86±0.51 6.47±0.96 6.53 9.45±1.60 1.66 3.80 1.00 0.37 5.21±2.50 1.97±0.16 10.8 24.9±3.6 12.9±1.48 24.1±1.56 12.0 4.35±5.33 18.6±137 0.20 1.50±4.93 2.28±0.24 12.7±1.57 43.6±11.6 21.9±2.31 73.4 177 6.24±0.45 <0 0.11 1.14±2.00 13 Table S2-7. Rate equations for Model v r1 r2 r3 r4 r5 r6 1 K 1 K k s1 K gly C gly K OH COH C gly K EG C EG 1 K OH COH 2 gly k s 2 K gly C gly K OH COH C gly K EG C EG 1 K OH COH 2 gly k s 3 K1, 2 PDO C1, 2 PDO K OH COH 1 K 1 K 1 K 1 K C1, 2 PDO 2 1, 2 PDO k s 4 K gly C gly K OH COH C gly K EG C EG 1 K OH COH 2 gly k s 5 K gly C gly K OH COH C gly K EG C EG 1 K OH COH 2 gly k s 5 K EG C EG K OH COH C gly K EG C EG 1 K OH COH 2 gly Table S2-8. Parameter estimation for Model v Model v Constants ks1 ks2×101 ks3×10-1 ks4 ks5 ks6 Kgly×10-1 KOH-×10-2 K1,2-PDO KEG×10-3 130oC 145oC 160oC 25.2±1.74 1.15±0.09 5.52±1.30 1.37±0.17 <0 8.59 1.55 1.40 8.30±7.07 1.08 4.12±2.47 0.23±0.14 7.03 0.27±0.17 0.51±0.31 0.03±0.04 0.77 47.8±18.2 0.01±0.005 4122±8719 - 14 Table S2-9. Rate equations for Model vi r1 r2 r3 1 K 1 K k s1 K gly C gly COH C gly 1 K OH C OH 2 gly k s 2 K gly C gly COH C gly 1 K OH COH 2 gly k s 3 K1, 2PDO C1, 2PDO COH 1 K r4 r5 r6 1 K 1 K 1 K C1, 2PDO 2 1, 2 PDO k s 4 K gly C gly COH C gly 1 K OH COH 2 gly k s 5 K gly C gly COH C gly 1 K OH COH 2 gly k s 5 K gly C gly COH C gly 1 K OH COH 2 gly Table S2-10. Parameter estimation for Model vi Model vi Constants ks1 ks2×101 ks3×10-2 ks4×10-2 ks5×10-2 ks6×10-2 Kgly×10-2 KOH-×10-3 K1,2-PDO 130oC 145oC 160oC 1.28±0.10 5.86±0.51 6.47±0.01 6.52 9.45±1.63 3.41±1.63 3.80 1.00 0.37 1.98±0.16 1.08 24.9±3.60 12.9±1.48 24.1±1.57 12.1±33.1 4.16±4.49 100.6±109 0.20 - 15 3. Error analysis Table S3-1. Repeated experiments in glycerol conversion on Pt/C catalyst T (oC) Time (h) Cgly CLA C1,2-PDO CEG CMeOH CEtOH CPrOH COH- 130 130 130 130 145 145 145 145 160 160 160 160 1.0 1.0 4.5 4.5 1.0 1.0 4.0 4.0 0.5 0.5 4.0 4.0 0.086 0.075 0.190 0.182 0.108 0.127 0.303 0.265 0.265 0.303 0.301 0.342 0.029 0.034 0.067 0.063 0.032 0.034 0.124 0.109 0.109 0.124 0.144 0.164 0.004 0.004 0.005 0.004 0.015 0.018 0.010 0.009 0.009 0.010 0.011 0.012 0.019 0.016 0.023 0.020 0.022 0.019 0.046 0.040 0.040 0.045 0.206 0.234 0.008 0.006 0.012 0.012 0.011 0.014 0.028 0.024 0.024 0.028 0.071 0.081 0.002 0.002 0.005 0.004 0.004 0.003 0.019 0.017 0.017 0.019 0.028 0.032 1.070 1.112 1.050 0.919 0.108 0.027 0.954 0.835 0.835 0.954 0.848 0.964 0.92 1.001 0.758 0.886 0.861 0.910 0.540 0.518 0.591 0.615 0.361 0.337 (1) Experimental error Key experiments in tandem glycerol conversion were repeated and values of substrate/product concentration were calculated (see Table S3-1), the error of which was taken into account when parameter estimation was carried out. (2) Measuring error The maximum error for HPLC analysis (repeated injection of one sample) is only < 0.08%, which is much lower compared with experimental error (approximately 3~12%). (3) Error analysis of reaction parameters-weighted least squares The goal of suing weighted least squares is to ensure that each data point has an appropriate level of influence on the final parameter estimation. However, it is difficult to evaluate the appropriate level of 16 each component (e.g. glycerol, lactic acid, etc), therefore the weight factor for each species and replicates (repeated experimental results) were considered is 1.0 during estimation of reaction parameters. (4) Error analysis in calculation of activation energy The activation energy was estimated based on reaction rate constants at each temperature. The uncertainty of activation energy for tandem glycerol conversion is summarized in Table 1-12. The equation for activation energy is k=ko×e(-Ea/RT). Table S3-2. Error analysis for activation energy in tandem glycerol conversion T (oC) Error in Ea (kJ/mol) 130 145 160 Ea ln(ko) (kJ/mol) ks1×100 1.33±0.09 1.97±0.15 4.09±0.59 53.0 23.82 0.958 0.42 0.76 0.30 0.49 ks2×10-1 6.09±0.51 10.8±0.50 22.8±3.04 63.7 26.24 0.991 0.53 5.38 1.57 1.88 ks3×10-2 6.48±0.98 24.8±3.38 55.6±14.0 104.2 36.19 0.985 5.57 13.22 5.07 7.14 ks4×10-2 5.95±0.53 12.9±1.48 39.3±4.23 104.2 35.81 0.967 1.55 13.30 4.47 5.25 ks5×10-2 7.00±1.87 24.2±1.64 131.6± 9.01 141.6 47.25 0.988 1.37 0.93 2.21 3.14 ks6×10-1 1.12±0.48 2.85±0.39 9.32±2.02 102.3 36.04 0.992 1.64 4.71 1.88 2.84 Kgly×10-2 3.83±0.47 3.75±0.25 3.50±0.48 4.3 3.21 0.902 -0.04 0.19 0.09 0.15 KOH-×10-3 2.21±0.17 1.81±0.22 1.33±0.29 24.5 -5.65 0.980 -0.32 0.77 0.28 0.41 K1,2-PDO×10-1 3.72±0.08 2.02±0.45 1.01±0.48 63.0 -12.01 0.997 1.80 6.82 2.40 3.19 KEG×10-2 4.00±0.37 2.50±0.48 1.13±0.12 60.9 -15.89 0.972 1.25 0.51 0.55 0.86 ki R2 err err (130) (145) err (160) err The unit for error is kJ/mol Error analysis in activation energy (example) (a) Functions for error propagation are listed in Table 1-13, which will be used to calculate the error (uncertainty) of activation energy. 17 Table 3-3. Functions for propagation of error Function Propagated error 𝑧 =𝑎+𝑏 ∆𝑧 = [(∆𝑎)2 + (∆𝑏)2 ]1/2 𝑧 =𝑎×𝑏 ∆𝑧 ∆𝑎 2 ∆𝑏 2 = [( ) + ( ) ] 𝑧 𝑎 𝑏 𝑧 = 𝑙𝑛𝑎 ∆𝑧 = 1/2 ∆𝑎 𝑎 ∆𝑎 is the uncertainty of 𝑎. (b) The uncertainty for activation energy is determined by the following equation: ln(𝑘 𝑇 ) = ln(𝑘𝑜 ) − 𝐸𝑎⁄ 𝑅𝑇 After rearrangement, 𝐸𝑎 = 𝑅𝑇[ln(𝑘𝑜 ) − ln(𝑘 𝑇 )] Therefore, the uncertainty for activation energy, 2 1/2 ∆𝐸𝑎 ∆𝑅𝑇 2 ∆[ln(𝑘𝑜 ) − ln(𝑘 𝑇 )] = [( ) +( ) ] [ln(𝑘𝑜 ) − ln(𝑘 𝑇 )] 𝐸𝑎 𝑅𝑇 2 1/2 ∆𝐸𝑎 ∆𝑇 2 ∆[ln(𝑘𝑜 )] + ∆ [ln(𝑘 𝑇 )] = [( ) + ( ) ] [ln(𝑘𝑜 ) − ln(𝑘 𝑇 )] 𝐸𝑎 𝑇 1/2 ∆𝐸𝑎 ∆𝑇 2 (∆𝑘𝑜 )/𝑘𝑜 + (∆𝑘 𝑇 )/𝑘 𝑇 2 = [( ) + ( ) ] [ln(𝑘𝑜 ) − ln(𝑘 𝑇 )] 𝐸𝑎 𝑇 To calculate the average error for 𝐸𝑎 based on different temperature, 18 2 2 1/2 2 ∆𝐸𝑎 = [(∆𝐸𝑎 𝑇1 ) + (∆𝐸𝑎 𝑇2 ) + (∆𝐸𝑎 𝑇2 ) ] (c) Take ks3 at 160 oC (433 K) for example. 2 1/2 ∆𝐸𝑎 𝑇1 (∆𝑘𝑜 )/𝑘𝑜 + (∆𝑘 𝑇1 )/𝑘𝑇1 ∆𝑇 2 = 𝐸𝑎 [( ) + ( ) ] 𝑇1 [ln(𝑘𝑜 ) − ln(𝑘 𝑇1 )] 1 13.9 0.9932 ) + = 98.9 1 2 2 (1 − 𝑘𝐽 1𝐾 2 49.9 × ( ) +( = 20.9 𝑘𝐽/𝑚𝑜𝑙 ) [34.58 − ln(49.9 × 10−2 )] 𝑚𝑜𝑙 433 𝐾 [ ] (d) Similarly, error at 130 oC and 145 oC is 1.11 kJ/mol and 2.44 kJ/mol respectively. (e) The average error for activation energy is calculated based on the following equation: ∆𝐸𝑎 𝑇1 + ∆𝐸𝑎 𝑇2 + ∆𝐸𝑎 𝑇3 2 ∆𝐸𝑎 2 ( ) =[ ] (3 × 𝐸𝑎 ) 𝐸𝑎 ∆𝐸𝑎 = ∆𝐸𝑎 𝑇1 + ∆𝐸𝑎 𝑇2 + ∆𝐸𝑎 𝑇3 1.11 + 2.44 + 20.9 = = 8.16 𝑘𝐽/𝑚𝑜𝑙 3 3 4. Sensitivity analysis (1) False compensation If reaction rates are measured over only a fairly small range of temperature and the activation energy is determined based on the reaction rate constants derived from Arrhenius plot, the value determined may be subject to considerable error. In other words, the sensitivity of obtained activation energy to temperature may be much more significant compared with the maximum error from reaction constants. Therefore, it is necessary to analyze the sensitivity of activation energy derived from kinetic modeling. 19 The sensitivity of activation energy is calculated based on the temperature range and error of reaction constants within the investigated temperature. The general equation is 𝛿𝐸𝑎 = 2 × 𝑅 × 𝑇1 × 𝑇2 × 𝑒𝑟𝑟𝑚𝑎𝑥 % |𝑇1 − 𝑇2 | Where 𝛿𝐸𝑎 is maximum error (sensitivity) of activation energy (kJ/mol), 𝑅 is the gas constant (kJ/mol.K), 𝑇1 and 𝑇2 are investigated temperature (K) and 𝑒𝑟𝑟𝑚𝑎𝑥 % is the maximum error in reaction constants (determined from 95% confidence level from Table S3-2). (2) Sensitivity of activation energy Table S4-1. Sensitivity of activation energy in tandem glycerol conversion relative error at T (oC) ki 130 ks1 ks2 ks3 ks4 ks5 ks6 Kgly KOHK1,2-PDO KEG 0.07 0.08 0.15 0.09 0.27 0.43 0.12 0.08 0.02 0.09 145 160 0.08 0.14 0.05 0.13 0.14 0.25 0.11 0.11 0.07 0.09 0.14 0.22 0.04 0.14 0.12 0.26 0.22 0.48 0.19 0.11 Maximum error 𝛿𝐸𝑎 (kJ/mol) 14.0 12.9 24.4 11.1 25.8 41.5 13.3 25.3 46.0 18.6 For example, the maximum error for ks1 is 0.14 in tandem glycerol conversion (see Table 1-12), while the temperature difference is 433 K (160 oC)-403 K (130 oC) = 30 K. Therefore the sensitivity of Ea for ks1 is 𝛿𝐸𝑎 = 2 × 8.314 × 433 × 403 × 0.14 = 14.0 𝑘𝐽/𝑚𝑜𝑙 1000 × 30 20 Evaluation of external and internal mass transfer limitation Intraparticle transfer limitation of glycerol: d m 1 p R initial p 6 2 D e w cat C gly 0.5 d p 0.0001m p 2.3 103 kg / m 3 R initial 0.52kmol / m 3 .h (maximum reaction rate at 160 oC) D e 9.4 10 -9 cm 2 / s 0.34m 2 / h (data at 25 oC) w cat 9.9kg / m 3 (maximum catalyst loading) C gly 1.1kmol / m 3 0.000299 Even if the error of parameter is 10000%, the significance intraparticle mass transfer limitation (0.0299) is still much lower than 0.2. Effect of stirring rate: (See Table 2 for experimental conditions, reaction time 2 h) At 100 RPM: glycerol conversion is 3.6% (indicating negligible conversion during startup). 21 At 500 RPM: glycerol conversion is 42.1%. At 800 RPM: glycerol conversion is 44.9%. 22