NMR
advertisement

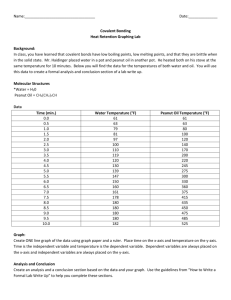
Matthew Marthaler March 16, 2012 Instrumental Lab Laboratory 6: FT-NMR Laboratory Introduction: In this lab, I will be working with Fourier Transform Nuclear Magnetic Resonance (FT-NMR) and becoming more familiar with its uses and techniques. NMR measures the absorption of electromagnetic radiation in the radio-frequency region of roughly 4 to 400 MHz. The nuclei of atoms rather than the electrons are involved in the absorption process. In FT-NMR, nuclei in a strong magnetic field are subjected periodically to very brief pulses of intense RF radiation whose length is usually less than 10 us with a frequency of 102 to 103 MHz. The free-induction decay (FID) signal is excited by the excited nuclei as they relax. The FID signal is digitalized and stored in a computer for data processing. The data is converted into a frequency domain signal by a Fourier Transformation and digital filtering may be applied to the data to further increase the signal-to-noise ratio. In easier terms, the sample is placed in a strong magnetic field and the nuclei affected by the field can be investigated. Purpose: The ultimate purpose of this procedure is to determine the structures of organic compounds. For the first day, we will be running standard and unknown organic compounds on the FT-NMR. We will also start proton (measures H’s) and C13 (measures carbons) of samples. For day two, we will finish proton and C13. We will then run a 2D analysis of the samples. Procedure: Day One 1) Make standards from Crisco pure vegetable oil, Planters peanut oil, Pompeian extra virgin olive oil, Stearic acid, and Palmitic acid. 2) The make the samples of the three oils, place 4 drops of the oil in an NMR tube and then put chloroform in the tube until the liquid reaches 4 – 4.5 cm. 3) To make the acid samples, place 0.08 g in an NMR tube and then put chloroform in the tube until the liquid reaches 4 – 4.5 cm. 4) Run standard samples on the NMR (S.O.P in my notebook). 5) Pick unknown A or B and run it. 6) Begin Proton and C13 of the samples. Day Two 1) 2) 3) 4) Run standard samples on the NMR. Pick unknown A and run it. Do proton and C13 analysis on the samples. Run 2D analysis of the samples. Results: Chemical Structures of the Standards Stearic Acid Palmitic Acid Crisco Pure Vegetable Oil + oil = Planters Peanut Oil + oil = Pompeian Extra Virgin Olive Oil + oil = 1H Proton Analysis for Our Standards and Unknowns Stearic Acid Chemical Shift Integration 0.9 1.278 1.654 2.366 35.85 35.85 35.85 2.30 Carbon Chain - 5 Planters Peanut Oil Chemical Shift Integration 0.897 1.276 1.617 1.638 2.044 2.089 2.319 2.777 4.141 4.293 4.812 5.351 7.281 68.68 68.68 68.68 68.68 68.68 14.52 14.52 1.89 1.87 1.81 3.98 7.25 n/a Carbon Chain - 33 Crisco Pure Vegetable Oil Chemical Shift (ppm) 5.344 4.178 4.158 2.774 2.291 2.316 1.260 Integration 9.84 1.95 1.96 3.91 5.97 9.82 66.56 Carbon Chain - 52 Extra Virgin Olive Oil Chemical Shift (ppm) 5.358 4.815 4.319 4.162 2.777 2.320 1.309 Integration 6.16 2.33 1.80 1.90 0.69 5.63 81.48 Carbon Chain - 35 Unknown A Chemical Shift Integration 0.889 1.276 1.385 1.639 1.736 2.046 2.321 2.778 4.143 4.280 4.816 5.331 7.281 65.99 65.99 65.99 65.99 65.99 9.35 5.73 3.25 1.87 1.86 3.09 8.86 n/a Carbon Chain - 39 From our calculations, I believe that the unknown is a mixture of Crisco vegetable oil and Planter’s peanut oil. This is, because the Carbon chain length is close to the number of Carbons in the peanut oil. Also, the integration numbers on the unknown came up to look like the integration numbers for Crisco vegetable oil and Planter’s peanut oil. Calculations Percent errors for Carbon Length: Stearic Acid: Peanut Oil: 5−18 18 33−30 30 Vegetable Oil: Olive Oil: x 100 = 72.22% error 52−51 35−18 18 x 100% = 10% error 51 x 100% = 1.96% error x 100 = 94.44% error Conclusion In conclusion, we were able to run both C13 APT and 1H Proton analysis on our standards and compounds, but we were not able to run a 2D analysis (not enough time). We made the samples by putting the desired amounts of chloroform and our sample/standard directly into the NMR tube, but later found out this was not the way to prepare samples (there was no mention of how to prepare the samples in the S.O.P though and we did not know this was an inaccurate way of making them). We then tested stearic acid and Palmitic acid first, followed by vegetable, peanut, and olive oil, and then we ran our unknown. We ran out of time to get a 1H Proton analysis for Palmitic acid, but that was the only sample we missed. Our results came out adequately, but the 1H Proton spectra for the different oils came out with numerous chemical shift values. This could be due to the oils having numerous signals, our improper way of making the samples, improper shimming, or to excess noise. We were still able to read the chemical shift and integration numbers for our standards and samples though, even with this problem. We calculated the percent error for the Carbon lengths and some came out well and others did not. This could be due to the fact that some samples were made better than others (less chloroform, mixed better, correct amount of sample), the instrument works better for different samples, or the noise was better for different samples. From our calculations, I believe that the unknown is a mixture of Crisco vegetable oil and Planter’s peanut oil. This is, because the Carbon chain length is close to the number of Carbons in the peanut oil. Also, the integration numbers on the unknown came up to look like the integration numbers for Crisco vegetable oil and Planter’s peanut oil. If anything could be changed to the experiment, it would be to have a proper explanation of how to make the samples and have the instruction manual or experts give a brief review on how to interpret the data. Overall this lab was more frustrating than others (because of the long time to run each sample and it coming up with poor results), but it still helped me gain a grasp on how to run the NMR.