Massep-GBMSDG sympos..
advertisement

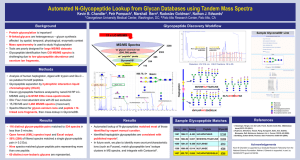
Confident assignment of site-specific glycosylation using a LC-MS/MS methodology with online HILIC enrichment Kshitij Khatri1, Gregory O. Staples2, Nancy Leymarie1, Deborah R. Leon1, Lilla Turiák1, Yu Huang1, Christian F. Heckendorf1 and Joseph Zaia1 1 Center for Biomedical Mass Spectrometry, Department of Biochemistry, Boston University School of Medicine, Boston, MA, USA 2 Agilent Technologies, Santa Clara, CA, USA Introduction: Glycans that decorate surface of proteins are critical for their biological functions. These glycans are highly heterogeneous and any changes in their composition and structure can alter the protein’s interaction with protein lectin domains that mediate linked biological processes. It is therefore important to characterize, compare and understand the role of these carbohydrate moieties in a sitespecific manner, to shed light on potential biomolecular interactions and to facilitate therapeutic interventions. Currently, the field lacks a workflow for accurate and reproducible analysis of site-specific protein glycosylation. Non-glycosylated peptides in glycoprotein digests, owing to their higher ionization efficiencies, adversely affect glycopeptide analysis by LC-MS (liquid chromatography-mass spectrometry). Therefore, glycopeptides need to be enriched prior to any LC-MS analyses. Here, we present a methodology for confidently assigning glycopeptides by combining online glycopeptide enrichment and separation with tandem MS. Methods: Standard glycoproteins were purchased from Sigma-Aldrich. Purified viral Hemagglutinin samples were purchased from Immune Technology, New York, NY. Glycoproteins were reduced, alkylated and subjected to tryptic digestion. Desalted glycoprotein digests were subjected to LC-MS/MS on an Agilent 6550 Q-TOF mass spectrometer coupled with Agilent 1200 series nanoflow HPLC system and Chip-cube nanospray source. A custom-made HPLC-chip separation device, with HILIC (Hydrophilic interaction liquid chromatography) enrichment column and reversed-phase analytical column was used for online enrichment and separation of glycopeptides. Whole tryptic digests were injected onto the HILIC column in high organic-solvent conditions to allow glycopeptide retention, while non-glycosylated peptides were washed away. The trapped glycopeptides were eluted from the HILIC column and resolved on the reversed-phase analytical column using a valve-switch and introduction of high-aqueous mobile phase followed by a reversed-phase gradient. Glycopeptides eluting were introduced into the mass spectrometer using an integrated nano-sprayer as they eluted from the analytical column and analyzed using CID (Collision-induced dissociation) tandem MS. LC-tandem-MS data were analyzed both manually and using a custom software tool, developed in-house, for analysis of large glycopeptide LC-MS/MS datasets. Results: Because glycosylation presents as a very heterogeneous modification, it can lead to a large search space when profiling glycopeptide LC-MS data. This large search space arising from different combinations of glycans and peptides can lead to ambiguous or false assignments if only MS1 data is used. The use of tandem MS therefore becomes imperative for confident assignment of glycopeptides. Since complexity in glycopeptide data ranges with number of glycosylation sites present on a glycoprotein, we demonstrated the usefulness of our method using glycoproteins ranging in number of glycosylation sites. Human transferrin (2 glycosylation sites), alpha-1-acid glycoprotein (5 glycosylation sites) and hemagglutinin from influenza-A-virus (9 glycosylation sites) were analyzed using typical C18 and HILICC18 chromatography with tandem-MS. The HILIC-C18 chromatography takes advantage of HILIC to trap glycosylated peptides by the virtue of their hydrophilic glycan moiety and depletes any non-glycosylated peptides from the sample. Because the resolution power of HILIC for glycopeptides is limited, we combined a HILIC enrichment column with reversed-phase analytical column for efficient separation of enriched glycopeptides. When compared with a commercially available C18 chip using typical reversed-phase gradient conditions, the HILIC-C18 chip showed significant improvement in glycopeptide signal abundances thereby enhancing the ability to perform data-dependent tandem-MS on glycopeptides. Tandem-MS allowed unambiguous assignment of glycopeptides using features like oxonium ions, stub glycopeptides and most importantly, peptide backbone ions. Each of the listed features from tandem MS, adds confidence to the glycopeptide assignments. Our prototype software for analysis of glycopeptide tandem MS allowed rapid interpretation of MS2 spectra and scoring of glycopeptide assignments. Using our improved chromatography system with online glycopeptide enrichment and separation followed by tandem MS, we were able to get confident site-specific glycan profiles for each of the three glycoproteins studied.