Automated N-Glycopeptide Lookup from Glycan
advertisement
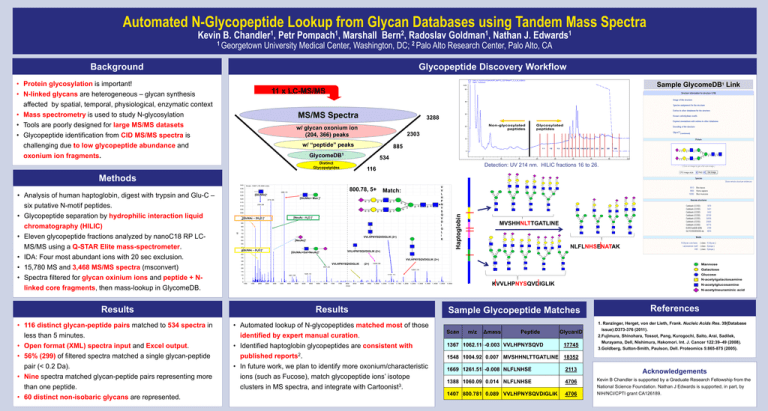
Automated N-Glycopeptide Lookup from Glycan Databases using Tandem Mass Spectra Kevin B. 1 Chandler , 1 Georgetown Petr 1 Pompach , 2 Bern , Marshall Radoslav 1 Goldman , Nathan J. 1 Edwards University Medical Center, Washington, DC; 2 Palo Alto Research Center, Palo Alto, CA Background Glycopeptide Discovery Workflow • Protein glycosylation is important! • N-linked glycans are heterogeneous – glycan synthesis affected by spatial, temporal, physiological, enzymatic context Sample GlycomeDB1 Link 11 x LC-MS/MS • Mass spectrometry is used to study N-glycosylation • Tools are poorly designed for large MS/MS datasets • Glycopeptide identification from CID MS/MS spectra is challenging due to low glycopeptide abundance and oxonium ion fragments. MS/MS Spectra 3288 w/ glycan oxonium ion (204, 366) peaks 2303 w/ “peptide” peaks 885 GlycomeDB1 534 Distinct Glycopetpides Detection: UV 214 nm. HILIC fractions 16 to 26. 116 • Analysis of human haptoglobin, digest with trypsin and Glu-C – six putative N-motif peptides. • Glycopeptide separation by hydrophilic interaction liquid chromatography (HILIC) • Eleven glycopeptide fractions analyzed by nanoC18 RP LCMS/MS using a Q-STAR Elite mass-spectrometer. • IDA: Four most abundant ions with 20 sec exclusion. • 15,780 MS and 3,468 MS/MS spectra (msconvert) • Spectra filtered for glycan oxinium ions and peptide + Nlinked core fragments, then mass-lookup in GlycomeDB. Results 800.78, 5+ [GlcNAc]+ Match: [GlcNAc+ Man ]+ [GlcNAc – 2H2O ]+ [NeuAc - H2O ]+ VVLHPNYSQVDIGLIK (2+) [NeuAc]+ [GlcNAc – H2O ]+ [GlcNAc+Gal+NeuAc]+ MVSHHNLTTGATLINE NLFLNHSENATAK VVLHPNYSQVDIGLIK (2+) VVLHPNYSQVDIGLIK (2+) VVLHPNYSQVDIGLIK (2+) KVVLHPNYSQVDIGLIK Results • 116 distinct glycan-peptide pairs matched to 534 spectra in less than 5 minutes. • Open format (XML) spectra input and Excel output. • Automated lookup of N-glycopeptides matched most of those identified by expert manual curation. • Identified haptoglobin glycopeptides are consistent with • 56% (299) of filtered spectra matched a single glycan-peptide pair (< 0.2 Da). • Nine spectra matched glycan-peptide pairs representing more than one peptide. published reports2. • In future work, we plan to identify more oxonium/characteristic ions (such as Fucose), match glycopeptide ions’ isotope clusters in MS spectra, and integrate with Cartoonist3. • 60 distinct non-isobaric glycans are represented. V V L H P N Y S Q V D I G L I K Haptoglobin Methods References Sample Glycopeptide Matches 1. Ranzinger, Herget, von der Lieth, Frank. Nucleic Acids Res. 39(Database Scan m/z Δmass Peptide 1367 1062.11 -0.003 VVLHPNYSQVD GlycanID 17745 issue):D373-376 (2011). 2.Fujimura, Shinohara, Tossot, Pang, Kurogochi, Saito, Arai, Sadilek, Murayama, Dell, Nishimura, Hakomori. Int. J. Cancer 122:39–49 (2008). 3.Goldberg, Sutton-Smith, Paulson, Dell. Proteomics 5:865-875 (2005). 1548 1004.92 0.007 MVSHHNLTTGATLINE 18352 1669 1261.51 -0.008 NLFLNHSE 2113 Acknowledgements 1388 1060.09 0.014 NLFLNHSE 4706 1407 800.781 0.089 VVLHPNYSQVDIGLIK 4706 Kevin B Chandler is supported by a Graduate Research Fellowship from the National Science Foundation. Nathan J Edwards is supported, in part, by NIH/NCI/CPTI grant CA126189.