N-Glycopeptide Identification from CID Tandem Mass
advertisement

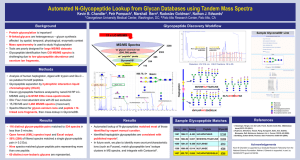
N-Glycopeptide Identification from CID Tandem Mass Spectra using Glycan Databases and False Discovery Rate Estimation Kevin B. Chandler, Petr Pompach, Radoslav Goldman, Nathan J. Edwards Georgetown University, Department of Biochemistry and Molecular & Cellular Biology, Washington, DC Introduction and Background Hypothesis and Aims • Over half of all proteins are glycosylated (this rate is higher for secreted, cell-surface and extracellular matrix proteins) • Glycosylation mediates cell-cell & cell-matrix interactions • N-linked glycosylation is enzyme-directed, occurs on Asn residues • We hypothesize that N-glycopeptide MS/MS data interpretation can be automated by adopting an algorithm that uses (1) oxonium marker ions and intact peptide peak filters, (2) the sequence(s) of the protein(s) of interest and (3) mass-matching to publicly available glycan databases to match glycan-peptide pairs to glycopeptide MS/MS spectra. within the motif NXS/T (X ≠ Pro) • Tandem mass spectrometry (MS/MS) is used to study protein glycosylation; however, there are few software tools to aid in processing and interpretation of large glycopeptide MS/MS datasets, and manual interpretation of datasets is time consuming • The aim of this research is to develop a novel software tool for rapid interpretation of glycopeptide MS/MS datasets to facilitate the study of glycoprotein microheterogeneity GlycoPeptideSearch Software – Glycopeptide Discovery Workflow GlycoPeptideSearch Scheme Methods: GlycoPeptideSearch Software 11 x LC-MS/MS MS/MS Spectra 3288 w/ glycan oxonium ion (204, 366) peaks 2887 w/ 2+ “peptide” peaks 317 GlycomeDB1 263 Distinct Glycopeptides 53 Test Dataset (Haptoglobin Glycopeptides) • Proteolytic digest of Haptoglobin with trypsin and GluC • Hydrophilic interaction liquid chromatography (HILIC) of glycopeptides • Eleven glycopeptide fractions analyzed by nanoC18 RP LC-MS/MS using a Q-STAR Elite mass-spectrometer. • IDA: Four most abundant ions with 20 sec exclusion. • 15,780 MS and 3,288 MS/MS spectra (msconvert) • Automated in silico digestion (Trypsin, GluC) of user-submitted protein sequence & N-glycosylation site ID • Fixed (carbamidomethylation) & variable modifications (Methionine oxidation) considered • Spectra filtered for glycan oxonium ions & peptide + N-linked core fragments & mass-lookup in GlycomeDB (glycan database) • Isotope Cluster Scoring performed on precursor ion • Decoy (non-motif containing) peptides submitted to search to enable estimation of the False Discovery Rate • Open format (XML) spectra input and Excel output. Summary of Haptoglobin Glycopeptides Controlling Error Using Spectra Filters and Hit Filters No. of Accepted Spectra Peptide Intensity Threshold 450 400 350 300 250 200 150 100 50 0 I=5 I=3 I=4 I=2 I=1 FDR Determination I = 10 I = 20 I = 30 0.0 1.0 2.0 3.0 4.0 5.0 6.0 Estimated FDR (%) 7.0 8.0 9.0 Target Peptides (with NXS/T) Decoy Peptides (without motif) Peptide Fragments (#) # Accepted Spectra 600 F=0 GPS 500 400 300 Sample MS/MS Spectrum: Fraction 17, Scan 1407 F=2 200 100 F=3 F=4 0 0 5 10 15 Estimated FDR (%) 20 25 Target and Decoy Spectra Hits # Accepted Spectra Isotope Cluster Score 350 300 250 200 150 100 50 0 IC = 50 IC = 20 IC = 10 IC = 5 IC = 2 IC = 1 0 2 4 IC = 100 IC = 200 6 8 10 Estimated FDR (%) IC = 9999 12 FDR 14 References Results and Conclusion 1. Ranzinger, Herget, von der Lieth, Frank. Nucleic Acids Res. 39(Database issue):D373-376 (2011). • 52 glycan-peptide pairs matched 263 spectra (3.9% FDR). • 52% (136) of filtered spectra matched a single glycopeptide pair (<0.2 Da), only 8 spectra matched 2. Fujimura, Shinohara, Tossot, Pang, Kurogochi, Saito, Arai, Sadilek, glycopeptide pairs representing > 1 peptide. • 27 distinct non-isobaric glycans at 4 sites were discovered, consistent with published reports2. 3. Goldberg, Sutton-Smith, Paulson, Dell. Proteomics 5:865-875 Murayama, Dell, Nishimura, Hakomori. Int. J. Cancer 122:39–49 (2008). (2005). 4. Pompach, Chandler, Lan, Edwards, Goldman. J.Proteome Res. 11 (3); 1728-40 (2012). Conclusion: Using characteristics of glycopeptide spectra including oxonium ions and intact peptide peaks, it is possible to automate glycopeptide CID MS/MS data interpretation with low false discovery. Acknowledgements Kevin B Chandler is supported by a Graduate Research Fellowship from the National Science Foundation. Nathan J Edwards is supported, in part, by NIH/NCI/CPTI grant CA126189. Rado slav Goldman is supported by NCI’s R01 CA115625 and CA135069.