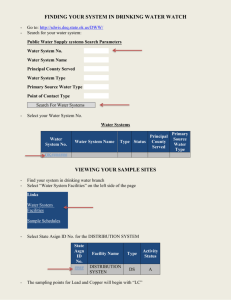
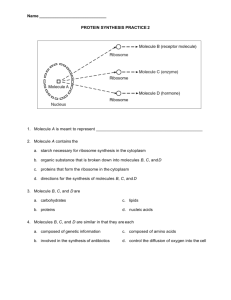
homework_problem_set_1
advertisement

MAE 6291
Fall 2012
Homework problem set 1
Binding
1. Suppose you can put 1012 antibodies on a 1 cm2 surface (1 antibody occupies ~ 10nm x 10nm of
surface; this is a rough estimate of the size of a protein) and you have a 1cm2 sensor surface in a
sensor that holds 0.1ml of sample volume. The fraction, f, of antibodies that bind analyte (the
molecule you are trying to measure), as a function of free analyte concentration [analyte], can
be estimated as f = [analyte]/KD / {1 + [analyte]/KD}. Note KD (the dissociation constant, koff /kon)
provides a measure of the tightness of binding since it is the concentration of (free) analyte at
which half the antibody molecules have bound analyte. The KD for antibodies is often nM. If
you can detect a signal over background when at least 1 in 1000 antibody molecules has bound
an analyte molecule (f = 10-3), what is the limit of sensitivity of the sensor in terms of number of
analyte molecules bound and concentration of free analyte molecules/sample volume? What if
you can detect when 1/106 or 1/109 antibodies has bound analyte? You might make a table with
columns “f”, “# analyte molecules bound”, and “[analyte]”.
Sensors are often limited by non-specific sticking to the sensor surface. E.g., if the sensor detects mass
or index of refraction change (mass), non-analyte proteins in the sample may stick and generate signal,
and 1 non-analyte molecule/106 antibody molecules (in a 10micron x 10micron area) might represent an
extremely “low-non-specific-binding” surface. If the sensor works by detecting a fluorescently labeled
antibody sticking to captured analyte, 1 molecule of fluorescent antibody sticking in the absence of
analyte (per 106 antibody molecules or 100 square microns of area) might provide a “floor” to the
minimum concentration detectable, as estimated above.
ELISA Sensitivity
2. The following figure is taken from
http://www.ebioscience.com/human-il-1-beta-high-sensitivity-elisa-kit.htm
describing a high-sensitivity ELISA for a protein involved in inflammation, IL-1.
Assume the absorption at analyte concentration 0 is 0.1, which would suggest non-specific sticking of
secondary antibody or conversion of substrate in the absence of enzyme. Estimate sensitivity in terms of
# molecules of analyte if analyte has MW 20,000 (i.e. 20,000g=1mol), you can reliably detect 1pg/ml =
1000pg/l = ? pM, and sample volume =100ul.
Assume capture and detection antibodies bind with nM affinity (KD = nM), and 2 x 1011 capture
antibodies in well (1012/cm2; well ~ 6mm diameter = .2cm2); what fraction of analyte do you expect to
bind to antibody? About how many enzyme molecules do you need in the well to get a signal above
background? If each enzyme molecule converts substrate to dye molecules at a rate of 1000/s, what is
the concentration of dye after 30minutes? How long would it take to get this concentration of dye
molecules if you could confine each captured enzyme molecule to a volume of 10-12l (this relevant to
next week’s paper)?
Brownian motion
3. Use the Einstein relation D = kBT and formula for the drag coefficient of a sphere r to
estimate the radii of the following molecules, assuming they are spheres, given their D values in
water at room temperature. If the radius of a virus is 100nm, what do you estimate for its
diffusion constant D?
Molecule
D (m2/s)
Water
2*10-9
Glucose
7*10-10
IgG
3*10-11
virus
r (10-9 m)
100
4. You want to observe the above virus infecting a cell, so you fluorescently label the virus and check
that you can see individual virus particles microscopically as tiny dots when a dilute sample is dried on a
slide, using a camera that collects light with 30ms exposure time. You wonder if the virus particles will
move too quickly in liquid to follow them using this camera. You reason that if a virus particle moves
<.5m in liquid (about a wavelength or diffraction-limited spot size) in 30ms, you should be able to
detect it. Will you be able to follow the virus? If your result suggests the virus might diffuse too quickly,
you could try adding glycerol to increase the viscosity of the cell culture medium. You think the cells will
tolerate glycerol up to 15% w/vol, which http://physics.mff.cuni.cz/vyuka/zfp/glyc_viscosity.pdf
suggests will raise the viscosity to about 1.5*10-3 Ns/m2 at room temperature. How much might this
help?
5. Using the formulas from the simple model of Brownian motion discussed in class, D = kT/6r =
2/2= 2mD/kBT, estimate m, D, and for a plastic bead 2m in diameter in water (assume bead
density = that of water, 1g/ml). Based on your estimates and the video of Brownian motion at
http://www.youtube.com/watch?v=iB7Eu7U73qg (assume the black spheres in the video are 2m
diameter beads), are the “jiggles” one sees individual and steps? If not, perhaps they are
displacements that arise from summing up the result of n steps, where 1/n is the camera rate in the
video or the rate your brain processes what you see. How many bead diameters would you expect a
bead to move in 1 sec?