bit25214-sm-0001-SuppData
advertisement

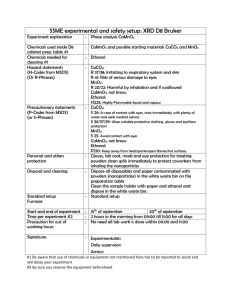
Supporting Information Directed Evolution of a Cellodextrin Transporter for Improved Biofuel Production under Anaerobic Conditions in Saccharomyces cerevisiae Jiazhang Lian1,2, Yanglin Li1,2, Mohammad Hamedi Rad1,2, Huimin Zhao1,2,3* Running Title: CDT Engineering via Directed Evolution 1 Department of Chemical and Biomolecular Engineering, 2 Energy Biosciences Institute, Institute for Genomic Biology, 3 Departments of Chemistry, Biochemistry, and Bioengineering, University of Illinois at Urbana- Champaign, Urbana, IL 61801 * To whom correspondence should be addressed. Phone: (217) 333-2631. Fax: (217) 333-5052. E-mail: zhao5@illinois.edu. 1 Supplementary Table S1. Oligonucleotides used in this study. Oligonucleotide CDT-F CDT-R Sequence (5’-3’) gtttcatttttcttgttctattacaac ctattattttagcgtaaaggatgggg Application Clone CDTs CDT-H-F1 CDT-H-R1 CDT-H-F2 CDT-H-R2 acgacgttgtaaaacgacggccagtgagcgcgcgtaatacgcaatgctactattttggag aattgatctatcgatttcaattcaattcaatctgcagtttgtaattaaaacttagattag aagcatagcaatctaatctaagttttaattacaaactgcagattgaattgaattgaaatc ctcactaaagggaacaaaagctggagctccaccgcggtgcaggaagaatacactatactg Construct CDT engineering helper plasmid CDT2-F CDT2-R attagaaagaaagcatagcaatctaatctaagttttaattacaaaatgggcatcttcaac gagaaaagaaaaaaattgatctatcgatttcaattcaattcaattcaagcaacagacttg CDT2 engineering primers CDT2-PM-R1 CDT2-PM-R2 CDT2-PM-F1 CDT2-PM-F2 CDT2-PM-F3 CDT2-PM-F4 CDT2-PM-R3 caaggcgggaatgcgccatgaccag gatgatggcgttgccggaccattgg tcaacaacgactggtcatggcgcattcccgccttgctccatgctttcccctccatcattc aacgactggtcatggcgcattcccgccttgctccaggctatcccctccatcattcagctc gtgggttcccgagtctccccgtttc caccgactccaccgccaaactcggtc ggcaacatcggcctcatcgccatcaatg Construct different combinations of sitedirected mutations CDT2-SM-F1 CDT2-SM-F2 CDT2-SM-F3 tacatcaacaacgactggtcatggcgcattcccgccttgctcNNSgctttcccctccatc ctcggcttcttctcccaatggtccggcaacgccatcatctccNNStactcctccaagctg cttgccaaggtcattgatggcgatgaggccgatgttgcccacNNSgacattcaccaggtc Construct saturation mutagenesis libraries at desired sites CDT2-eGFP-R1 CDT2-eGFP-F2 CDT2-eGFP-R2 gaacagctcctcgcccttgctcaccatactaccactaccagcaacagacttgccctcatg attcacgagcatgagggcaagtctgttgctggtagtggtagtatggtgagcaagggcgag aaagaaaaaaattgatctatcgatttcaattcaattcaatttacttgtacagctcgtcca Construct CDT2 & its mutants fused with eGFP 2 Supplementary Table S2. Comparison of cellobiose fermentation performance of CDT1, CDT2, and CDT2 mutants obtained in the first (HIH), second (HH), and third (HHT) round of directed evolution using high cell density fermentation. Ethanol used to solubilize anaerobic growth factors (0.5-1 g/L) was subtracted before calculating ethanol productivity and ethanol yield. To determine the cellobiose consumption rate and ethanol productivity, data points within the linear range were included for linear regression. Final data points were used to determine ethanol yield. Cellobiose Consumption Rate (g/L·h) Ethanol Productivity (g/L·h) Ethanol Yield (g/g cellobiose) WT HIH HH HHT CDT1 0.67±0.01 1.5±0.07 1.8±0.19 2.7±0.02 1.1±0.00 0.25±0.00 0.55±0.04 0.66±0.06 1.1±0.01 0.38±0.02 0.35±0.00 0.39±0.02 0.40±0.01 0.45±0.01 0.39±0.01 3 Supplementary Table S3. Saturation mutagenesis of CDT2 at the sites of Q207, N311, and I505. Saturation mutagenesis libraries were constructed using degenerate primers. After spreading into SCC-Leu plates, the top 12 colonies from each library were picked up and further verified in cellobiose liquid medium. The top 5 clones showed increased growth rates on cellobiose were chosen and the sequences at the desired sites were determined by DNA sequencing. As for the site at I505, none of the clones grew faster than I505T. To validate the constructed saturation mutagenesis library, 4 colonies were randomly picked up from the N311 library on non-selective plate (SCD-Leu). Compared with the screened clones, which showed highly enriched codon triplets encoding Glycine, the sequences from the non-selective plate were completely random. Clones Q207(CAG) N311(AAC) Q2 Q5 Q9 Q11 Q12 N1 N2 N3 N5 N9 Top Clones Nucleotide Amino Acid Change Change ACC T GCC A ACC T ACG T ACC T GGG G GGG G GGC G GGC G GGC G 4 Clones N-R1 N-R2 N-R3 N-R4 Random Clones Nucleotide Amino Acid Change Change CGG GCG CTG AAC R A L N Supplementary Figure S1. Vector map of the helper plasmid for CDT2 engineering, CDT-EngH. NCU00130 is the β-glucosidase from N. crassa, and the cellobiose utilization pathways are constructed by inserting CDTs or CDT2 library into the PstI site, which was flanked by TEF1 promoter and PGK1 terminator. 5 Supplementary Figure S2. A general scheme to engineer sugar transporters, using the colony size based screening method. CDT2 library was constructed by error-prone PCR and cloned into CDT-Eng-H by the DNA assembler method. After spreading into the selective cellobiose agar plates, 60-80 largest colonies were picked up and subject to first tier screening in culture tube. The top 12 mutants with either the highest cell density or specific growth rate were selected, whose plasmids were extracted and re-transformed into fresh yeast cells to eliminate host adaptation. After re-transformation, the selected mutants were subject to the second tier screening in shake flask under oxygen limited conditions. The top 3 mutants with the highest cellobiose consumption rate and ethanol production rate were selected for final characterization (the third tier screening) under anaerobic conditions. For easier operation, the first and second tier screening was carried out under aerobic conditions, and only the third tier screening was performed under anaerobic conditions. Directed evolution was carried out iteratively until no further improvement was observed 6 Supplementary Figure S3. Characterization of the role of Q207H/F209I/N311H (HIH) mutations in cellobiose fermentation performance by creating all possible combinations, including Q207H, F209I, N311H, Q207H/F209I (HI), Q207H/N311H (HH), and F209I/N311H (IH). Fermentation was carried out with an initial OD600 of 0.05 inoculated into fresh YPC media. The growth curves of N311H, Q209H, WT, and IH at 28h were enlarged for easier comparison of their growth rate. 7 Supplementary Figure S4. Anaerobic cellobiose fermentation profiles of CDT2 mutants obtained in the first (HIH), second (HH), and third (HHT) round of directed evolution using the library screening conditions in rich media. The specific growth rate (A), cellobiose consumption rate (B), and ethanol productivity (C) of these transporters were compared. Ethanol used to solubilize anaerobic growth factors (0.5-1 g/L) was subtracted before calculating the titer, productivity, and yield of ethanol. To determine the specific growth rate, cellobiose consumption rate and ethanol productivity, data points within linear range were included for analysis. Final data points were used to determine ethanol yield. 8 Supplementary Figure S5. Mutant analysis by site-directed mutagenesis. The role of Q207H/N311H/I505T (HHT) mutations in cellobiose fermentation performance were characterized by creating and comparing all possible combinations, including the single mutants (Q207H, N311H, and I505T) and double mutants (HH, Q207H/I505T and N311H/I505T). Fermentation was carried out with an initial OD600 of 0.05 inoculated into fresh YPC media. 9 Supplementary Figure S6. Cellobiose fermentation profiles of CDT2 on single copy plasmid (CEN/ARS Ori) and multiple copy plasmid (2µ Ori), respectively. The growth rate (A) and cellobiose consumption rate (B) were compared under oxygen-limited conditions. Fermentation was carried out with an initial OD600 of 0.05 inoculated into fresh YPC media. 10 Supplementary Figure S7. Aerobic cellobiose fermentation profiles of CDT1, CDT2, and CDT2 mutant (HHT). The growth rate (A) and cellobiose consumption rate (B) were compared. Fermentation was carried out with an initial OD600 of 0.05 inoculated into fresh YPC media. 11