bit25810-sup-0002-SupInfo-S1
advertisement

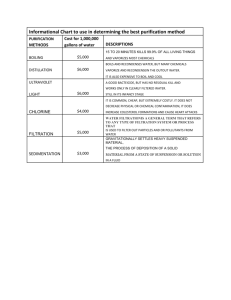
Experimental details of the Case Study: Low pH flocculation and Filtration An in-house comparative study assessed simultaneous clarification and purification by low pH flocculation in combination with depth filter screening for multiple antibodies. Impact on overall impurity removal in the harvest and Protein A purified material was investigated. As discussed in earlier sections of this review, depth filtration can also be used post Protein A chromatography to reduce levels of HCP and DNA. In this case study, impurity clearance with and without low pH flocculation and depth filters differing in charge density post Protein A were compared. As shown throughout, key responses such as impurity and viral clearance were assessed. Experimental Materials and Methods. The antibodies (mAb A and mAb B) used in this study were produced in a Chinese hamster ovary (CHO) cell line by Bristol-Myers Squibb. The cell culture fluid (CCF) producing mAb A (IgG4, pI ≈ 7.8) and mAb B (IgG4, pI ≈ 6.5) had cell densities of 1.4-1.8 x 107 cells/mL, The CCF had the following properties: viability at harvest of 50-70 %, antibody titer of approximately 3-3.5 g/L, an initial turbidity of approximately 3000 NTU. Product recovery for the harvest process was calculated based on product concentration before and after each step, which was determined using a Poros® Protein A HPLC column. HCP leves were quantified using an enzyme-linked immunosorbent assay (ELISA kit, Part #: F015, Cygnus Technologies, Southport, NC). A quantitative polymerase chain reaction (qPCR) method was used to quantitate residual DNA. The qPCR method used Taqman® chemistry with primers and a probe specific to CHO genomic DNA. The quantity of CHO DNA in the sample was calculated based on a standard curve generated using CHO genomic DNA as a template in the qPCR reaction. Size Exclusion HPLC (SEC) was used for the quantitative analysis of monomer, High Molecular Weight (HMW), and Low Molecular Weight (LMW) using a Tosoh Bioscience G3000 SWXL column (Part #: 08541, King of Prussia, PA). Residual dextran sulfate was determined using a high molecular dextran sulfate ELISA kit (Product #: K-4100, Lifespan Technologies, Salt Lake City, UT). 1 Flocculation and Depth filtration. The harvest treatment included low pH only and low pH adjustment followed by addition of the flocculant, which was a sodium salt of dextran sulfate (DS) with a molecular weight of 500 kDa (Sigma Aldrich). The conditions for flocculation were based on previous screening studies targeting recovery of ≥ 90% at acceptable levels of impurity clearance. The pH was adjusted by gradually adding 1M citric acid into the CCF to reach pH values of 6.0, 5.5, 5.0, and 4.5. Then, the DS stock solution (10 g/L in water) was added into the pH adjusted CCF to final concentrations of 0, 0.05, 0.1, and 0.5 g/L, respectively. After mixing for 60 minutes, all treated CCF were centrifuged at 500 x g for 10 minutes, followed by 0.2 µm sterilizing filtration using syringe filters. The harvested cell culture fluid (HCCF) treated with the optimized conditions (pH only and pH with DS) were then purified by affinity chromatography using a Protein A column (MabSelect SuRE, GE Healthcare, Piscataway, NJ). Both untreated and treated cell cultures were clarified using depth filtration for processing the clarified bulk for downstream purification. All the HCCF that were purified by affinity chromatography (Protein A) and the Protein A elution pools (referred to ‘eluate’ henceforth) containing the purified mAbs were further filtered comparing two depth filters with different levels of anionic charges (90ZB05A and Emphaze, Table-3). HCP breakthrough curves were generated for depth filters as shown in Figure S-1. The HCCF samples from the primary clarification (depth filtration), Protein A eluates and depth filtration post Protein A were analyzed for HCP, DNA and aggregates. The protein concentration of HCCF was also analyzed to determine the harvest recovery. 2