Section 10
advertisement

STAT 405 - BIOSTATISTICS
Handout 10 – Comparing Two Binomial Proportions with
Independent Samples
This handout covers material found in Sections 10.2 and 10.3 of your text.
EXAMPLE: Cardiovascular Disease (Examaple 10.6 of your text, page 389).
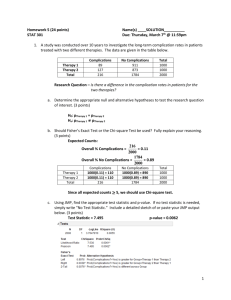
A study looked at the effects of oral contraceptive (OC) use on heart disease in women 40 to 44
years of age. It found that among 5,000 current OC users at baseline, 13 women develop a
myocardial infarction (MI) over a 3-year period. Among 10,000 non-OC users, 7 develop an MI
over a 3-year period. Assess the statistical significance of the results.
NORMAL-THEORY METHOD
In your introductory statistics course, you more than likely discussed the following hypothesis
testing procedure for comparing two proportions:
Check
Assumptions:
For this particular test, we make the following assumptions:
Set up the
Hypotheses:
n1 p̂1 ≥ 5, n1(1- p̂1 ) ≥ 5, n2 p̂ 2 ≥ 5, n2(1- p̂ 2 ) ≥ 5.
We assume the observations are independent.
Ho: The proportion of current OC users who develop an MI over a 3-year
period is equal to the proportion of non-OC users who develop an MI over
a 3-year period.
Ha: The proportion of current OC users who develop an MI over a 3-year
period differs from the proportion of non-OC users who develop an MI
over a 3-year period.
Calculate the
Test Statistic:
First, p
n 1 pˆ 1 n 2 pˆ 2
=
n1 n 2
Then,
𝑧=
(𝑝
̂1 − 𝑝
̂)
2
1
1
𝑛1 + 𝑛2 )
√𝑝̅ 𝑞̅ (
1
Find the
p-value:
data normalprob;
CumProb=CDF('Normal',-3.01,0,1); output;
proc print;
run;
p-value =
In addition, you calculated the confidence interval for the difference in proportions as follows:
Estimate the difference in proportions: (𝑝
̂1 − 𝑝
̂)=
2
Find the standard error for the estimate:
Find the appropriate percentile from the z-distribution:
Construct the confidence interval:
p̂1 (1 p̂1 ) p̂ 2 (1 p̂ 2 )
=
n1
n2
2
Constructing the Confidence Interval in SAS
data nutrition;
input OC$ MI$ count;
datalines;
User
Yes 13
User
No 4987
Nonuser
Yes 7
Nonuser
No
9993
;
proc freq order=data;
tables OC*MI / riskdiff;
weight count;
run;
3
THE CHI-SQUARE TEST FOR HOMOGENEITY OF BINOMIAL PROPORTIONS
This same hypothesis test can also be approached from a different perspective. Note that we
have two independent samples of women with different contraceptive-use patterns, and we want
to compare the proportion of women in each group who develop an MI. In this situation, one
set of margins in the contingency table are fixed (i.e., the row totals) and the number of
successes in each row is a random variable.
MI status over 3 years
Yes
No
Total
OC users
13
4987
5,000
Non-OC users
7
9993
10,000
Total
20
14,980
15,000
The test statistic is given as follows:
Test Statistic =
(Observed- Expected)2
Expected
To calculate this test statistic, we need to first find the counts that would be EXPECTED if there
were no difference in the proportion which developed an MI over 3 years between the OC-users
and non-users.
The Observed Counts
The Expected Counts
4
We can use SAS to calculate the expected counts and also the test statistic:
proc freq order=data;
tables OC*MI / riskdiff all expected nopercent nocol norow;
weight count;
run;
Verify the Chi-square test statistic:
χ2
(Observed- Expected)2
=
Expected
Question: What does it mean if the test statistic is “large”?
5
It can be shown that under the null hypothesis, the test statistic APPROXIMATELY follows a
chi-square distribution with 1 df. The p-value is calculated as follows:
data ChiSquareprob;
CumProb=1-CDF('ChiSquare',9.037,1); output;
proc print;
run;
General Comments:
1. Note that the p-value is the same as that provided using normal-theory methods.
2. The Chi-square test is ALWAYS a two-sided test! In contrast, one-sided tests can be
carried out using normal theory methods.
3. The Chi-square test of homogeneity should be used only when the normal approximation
to the binomial is valid. This can be shown to be approximately true if no expected count
in the table is less than 5.
6
Yates-Corrected Chi-Square Test
Under certain circumstances, a version of the chi-square test statistic with a continuity
correction yields more accurate p-values than does the uncorrected version when approximated
by a chi-square distribution. For the continuity-corrected version, the statistic is calculated as
follows:
χ2
1
Observed- Expected
2
Expected
2
Again, under the null hypothesis, the test statistic APPROXIMATELY follows a chi-square
distribution with 1 df.
Find the continuity-corrected test statistic:
2
χ2
1
Observed- Expected
2
=
Expected
Note that SAS PROC FREQ also returns this statistic and the associated p-value:
7
FISHER’S EXACT TEST
We have just discussed two tests (the Z-test based on normal theory methods and the ChiSquare test) which can be used for comparing two binomial proportions with independent
samples. However, both of these methods require that the normal approximation to the
binomial distribution be valid. This is often not the case, especially with small samples;
therefore, we often turn to Fisher’s exact test.
EXAMPLE: Cardiovascular Disease (Modified from Example 10.16 of your text, page 402).
Suppose a retrospective study is done among men aged 50-54 in a specific county who died over
a one-month period. The investigators try to include approximately an equal number of men
who died from CVD (the cases) and men who died from other causes (the controls). Of 15
people who died from CVD, 4 were on a high-salt diet before they died. In contrast, of 15 people
who died from other causes, 1 was on a high-salt diet. The data are shown in the following
contingency table:
High-Salt Low-Salt Total
Non-CVD
1
14
15
CVD
4
11
15
Total
5
25
30
Question of Interest: Is the proportion of the CVD group which has a high-salt diet greater than
the proportion of the Non-CVD group which has a high-salt diet, or is this observed difference
simply due to chance?
As always, we will find the probability of obtaining a sample at least as extreme as the observed
(assuming that there is no difference in risk between the two groups).
To calculate this probability, we need only consider that we have 5 high-salt dieters and 25 lowsalt dieters. Furthermore, assume that a high-salt dieter is equally likely to be from either group
(that is, there is no difference in the two proportions of interest). If we randomly divide these 30
individuals into two groups of size 15, what is the probability that four or more of the high-salt
dieters will end up in the CVD group? To find this probability, we will use the hypergeometric
probability distribution.
Hypergeometric Distribution
Assume that both the row and column totals of our contingency table are fixed. The number of
high-salt dieters in the CVD group (X) is a hypergeometric random variable. Since we are
assuming the column totals are fixed (so there are five high-salt dieters overall), this random
variable can assume the following values: x = 0, 1, 2, 3, 4, or 5.
The probability attached to each value can be calculated using the hypergeometric probability
distribution. Suppose a population consists of N items, k of which are “successes.” A random
sample drawn from that population consists of n items. The probability that, of these n items in
the sample, x are “successes” is given by:
8
k N k
x n x
P(X = x) =
for x = 0, 1, 2, … n.
N
n
In our example, recall that X = the number of high-salt dieters who died from CVD. We will
consider our “sample” to consist of the CVD group. Also, we will consider a “success” as having
a high-salt diet. Then, the notation is as follows:
N = total number in the study =
k = total number with high-salt diet =
n = total number in CVD group =
To find the probability of observing 4 high-salt dieters in the CVD group:
P(X = 4) =
In SAS:
data HypergeometricProb;
prob = pdf('hyper', 4, 30, 5, 15); output;
proc print noobs; run;
* In general, use pdf('hyper', x, N, k, n);
To find the probability of observing 5 high-salt dieters in the CVD group:
data HypergeometricProb;
prob = pdf('Hyper',5,30,5,15); output;
proc print noobs; run;
Finally, recall that we observed 4 high-salt diets in the CVD group. Therefore, the probability of
obtaining a sample at least as extreme as the observed is given by:
P(X = 4) + P(X = 5) =
9
Questions:
1. Does observing a sample at least as extreme as ours seem likely to happen by random
chance alone? Explain.
2. What conclusions can you draw concerning the research question?
Finding the p-value in SAS PROC FREQ
data HighSalt;
input COD$ Diet$ count;
datalines;
CVD HighSalt 4
CVD LowSalt 11
NonCVD HighSalt 1
NonCVD LowSalt
14
;
proc freq order=data;
tables COD*Diet / all;
weight count;
run;
10
Carrying out Fisher’s Exact Test
This test is based on the probability of observing a table at least as extreme as the original data.
The procedure is carried out as follows.
Step 1:
Convert the research question into H0 and Ha.
H0: The proportion of the CVD group which has a high-salt diet is equal to the
proportion of the Non-CVD group which has a high-salt diet.
Ha: The proportion of the CVD group which has a high-salt diet is greater than
the proportion of the Non-CVD group which has a high-salt diet.
Step 2:
Determine α, the error rate.
Step 3:
Determine the p-value and make a decision concerning H0.
p-value:
Decision:
Step 4:
Write a conclusion in terms of the original research question.
11
In JMP
First enter these data into a data table in JMP as shown below:
Next we use Fisher’s Exact test to compare the proportions with high salt in the case and control
groups. Because this is a case-control study the response is presence of the risk factor, high salt
in this case. The correct dialog box for Analyze > Fit Y by X is shown below.
The output from JMP is shown below:
12
EXAMPLE: Recall the following example from Handout 7.
A case-control study was conducted to determine whether there was an increased risk of cervical
cancer amongst women who had their first child before age 25. A sample of 49 women with
cervical cancer was taken of which 42 had their first child before the age of 25. From a sample of
317 “similar” women without cervical cancer it was found that 203 of them had their first child
before age 25.
Age ≤ 25
Age > 25
Column Totals
Cancer (Case) No cancer (Control) Row Totals
42
203
245
7
114
121
49
317
366
Again, we will find the probability of obtaining a sample at least as extreme as the observed by
random chance alone.
Questions:
1. What would the “most extreme” table look like? Note that we must keep the row and
column totals the same.
2. Give some examples of tables that are more extreme than the observed data but not as
extreme as the “most extreme” table.
13
We can find the probability of observing each of the following tables by once again using the
hypergeometric distribution. Let the random variable X = the number of women in the Cancer
group who were 25 or younger when they had their first child.
N = total number in the study =
k = total number who were 25 or younger =
n = total number in Cancer group =
X
Contingency Table
P(X=x)
x = 42
x = 43
x = 44
x = 45
x = 46
x = 47
14
x = 48
x = 49
Questions:
What is the probability of observing a table at least as extreme as ours by random chance
alone? What does this say about our research question?
Note that this test can also be carried out in SAS:
data CervicalCancer;
input Age$ Cancer$ Count;
datalines;
LE25 Case
42
LE25 Control 203
GT25 Case
7
GT25 Control 114
;
proc freq order=data;
tables Age*Cancer / all;
weight Count;
run;
15
In JMP
Analyze > Fit Y by X
Y = Preg. Age
X = Disease
Why is Disease the X variable?
Fisher’s Exact appears automatically for
2 x 2 tables. There is an additional option
you can select called Exact Test. This
will work for larger tables as well (slow!).
16
In R
> CervicalCancer = matrix(c(42,7,203,114),nrow=2,dimnames=
+ list(Cancer=c("Cervical","Control"),PregAge=c("Age < 25","Age > 25")))
Note: the default in R is enter matrices by column. If you want enter the matrix by rows rather than columns you
need to include the optional argument byrow=T in the matrix function call.
Cancer (Case) No cancer (Control) Row Totals
42
203
245
7
114
121
49
317
366
Age ≤ 25
Age > 25
Column Totals
Fisher's Exact Test for Count Data
fisher.test(CervicalCancer,alternative="two.sided")
data: CervicalCancer
p-value = 0.002937
∗ 𝐻𝑎 :
alternative hypothesis: true odds ratio is not equal to 1
∗∗ 𝐻𝑎 :
95 percent confidence interval: 1.433247 9.158597
sample estimates:
odds ratio
3.360159
Yate’s Corrected Chi-Square Test
prop.test(x=c(42,203),n=c(49,317))
2-sample test for equality of proportions with continuity correction
data: c(42, 203) out of c(49, 317)
X-squared = 8.0579, df = 1, p-value = 0.004531
alternative hypothesis: two.sided
95 percent confidence interval:
0.09367083 0.33985779
sample estimates:
prop 1
prop 2
0.8571429 0.6403785
OR
prop.test(CervicalCancer)
this will produce the same output as shown above.
17