tpj12469-sup-0010-Legends
advertisement

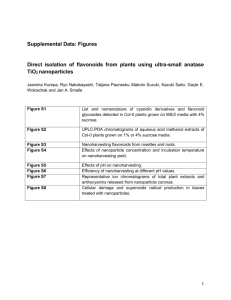
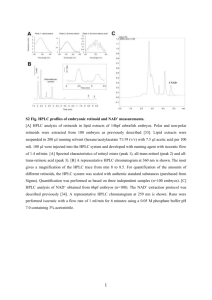
Supporting Legends Figure S1. Flower phenotypes of the efp mutants Ipomoea efp mutants and germinal revertants (a), and efp knockdown transformants of Petunia (b) and Torenia (c). The TT48 and TT49 series of Torenia transformants harbor the Torenia ThEFP-A and ThEFP-B RNAi constructs, respectively (Figure S5). Figure S2. Comparison of the EFP protein with its relatives (a) A phylogenetic tree for the CHI superfamily. The amino acid sequences were aligned with the ClustalW program of the MEGA 5 software (Tamura et al., 2011). The tree was constructed using the neighbor-joining method of the MEGA 5 with the Poisson correction model. Bootstrap values of 1000 replicates are shown next to the branches, and the scale bar indicates 0.1 amino acid substitutions per site. Accession numbers of the sequences are shown in parentheses, and the amino acid sequences of InCHI and InFAP were deduced from EST sequences. (b) Multiple alignments of the deduced amino acid sequences of Arabidopsis AtCHIL and EFP proteins. (c) Comparison of the catalytic residues of CHI with AtCHIL and EFP proteins. Sequences were aligned with the ClustalW program v2.1. Identical amino acids, highly similar residues, less similar residues, and gaps are indicated by asterisks (*), colons (:), periods (.), and hyphens (-), respectively. The conserved catalytic residues among CHI proteins are indicated by red characters (Jez et al., 2000; Ngaki et al., 2012). Abbreviations shown in front of each protein indicate the plant species: At, Arabidopsis thaliana; Gm, Glycine max; In, Ipomoea nil; Ms, Medicago sativa; Og, Oncidium cv. Gower Ramsey; Ph, Petunia hybrida; and Pp, Physcomitrella patens; and Th, Torenia hybrida. OgEFP, PpEFPa, PpEFPb, PpFAPb were originally called OgCHI, CHILa, CHILb, and FAPb, respectively (Chiou and Yeh, 2008; Ngaki et al., 2012). Chiou, C.Y. and Yeh, K.W. (2008) Differential expression of MYB gene (OgMYB1) determines color patterning in floral tissue of Oncidium Gower Ramsey. Plant Mol. Biol., 66, 379-388. Jez, J.M., Bowman, M.E., Dixon, R.A. and Noel, J.P. (2000) Structure and mechanism of the evolutionarily unique plant enzyme chalcone isomerase. Nat. Struct. Biol., 7, 786-791. Ngaki, M.N., Louie, G.V., Philippe, R.N., Manning, G., Pojer, F., Bowman, M.E., Li, L., Larsen, E., Wurtele, E.S. and Noel, J.P. (2012) Evolution of the chalcone-isomerase fold from fatty-acid binding to stereospecific catalysis. Nature, 485, 530-533. Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M. and Kumar, S. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular biology and evolution, 28, 2731-2739. Figure S3. HPLC analysis of anthocyanins in Ipomoea Anthocyanin structures (a) and HPLC analysis of anthocyanins in EFP (b) and efp-1(c) flowers. WBA (Wedding Bells Anthocyanin), pelargonidin 3-O-[2-O-(6-O-(trans-3-O-( β -D-glucopyranosyl)caffeoyl)β -D-glucopyranosyl)-6-O-(trans-4-O-(6-O-(trans-3-O-(β-D-glucopyranosyl)caffeoyl-β -D-glucopyranosyl)caffeoyl)-D-glucopyranoside]-5-O-[ β -D-glucopyranoside]; A1, pelargonidin 3-O-[2-O-(6-O-(trans-3-O-( β -D-glucopyranosyl)caffeoyl)- β -D-glucopyranosyl)- β -D-glucopyranoside]-5-O-[ β -D-glucopyranoside]; A2, pelargonidin 3-O-[2-O-(6-O-(trans-caffeoyl)β -D-glucopyranosyl)β -D-glucopyranoside]-5-O-[ β -D-glucopyranoside]; A3, pelargonidin 3-O-[2-O-(6-O-(trans-3-O-( β -D-glucopyranosyl)caffeoyl)β -D-glucopyranosyl)-6-O-(trans-caffeoyl)β -D-glucopyranoside]-5-O-[ β -D-glucopyranoside]. Absorbance at 530 nm was used for the detection of anthocyanin pigments. Figure S4. HPLC analysis of colorless flavonoids in Ipomoea HPLC analysis of colorless flavonoids in Ipomoea EFP (a) and efp-1 (b) flowers. Absorbance at 360 nm was used to detect colorless flavonoids. A1–3 are the anthocyanins presented in Figure S3a, and F1 is the flavonol kaempferol glycoside. C1–4 are caffeic acid derivatives: C1, chlorogenic acid; C2, 1-O-caffeoyl glucoside. Note that chalcone 2´-O-glucoside (retention time: 25.9 min), which is a major yellow flavonoid pigment that accumulates in CHI null mutants (Saito et al., 2011), was virtually absent in the efp-1 mutant. Saito, N., Tatsuzawa, F., Hoshino, A., Abe, Y., Ichimura, M., Yokoi, M., Toki, K., Morita, Y., Iida, S. and Honda, T. (2011) The anthocyanin pigmentation controlled by the speckled and c-1 mutations of the Japanese morning glory. J. Japan. Soc. Hort. Sci., 80, 452-460. Figure S5. RNAi vectors for Petunia and Torenia EFP knockdowns Restriction sites used for vector construction are indicated, and the sites within the parentheses are from cloning vectors. The restriction fragments were cloned into the pBINPLUS binary vector (van Engelen et al., 1995). MAC-1P and MAST are the Ti plasmid mannopine synthase promoter (Comai et al., 1990) and terminator, respectively. El235SP and NOST denote an enhanced cauliflower mosaic virus 35S promoter (Mitsuhara et al., 1996) and Agrobacterium tumefaciens nopaline synthase terminator, respectively. Comai, L., Moran, P. and Maslyar, D. (1990) Novel and useful properties of a chimeric plant promoter combining CaMV 35S and MAS elements. Plant Mol. Biol., 15, 373-381. Mitsuhara, I., Ugaki, M., Hirochika, H., Ohshima, M., Murakami, T., Gotoh, Y., Katayose, Y., Nakamura, S., Honkura, R., Nishimiya, S., Ueno, K., Mochizuki, A., Tanimoto, H., Tsugawa, H., Otsuki, Y. and Ohashi, Y. (1996) Efficient promoter cassettes for enhanced expression of foreign genes in dicotyledonous and monocotyledonous plants. Plant Cell Physiol., 37, 49-59. van Engelen, F.A., Molthoff, J.W., Conner, A.J., Nap, J.P., Pereira, A. and Stiekema, W.J. (1995) pBINPLUS: an improved plant transformation vector based on pBIN19. Transgenic Res., 4, 288-290.