GelElectrophoresisLab
advertisement

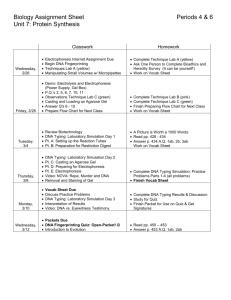
Name: _______________________________________ Date: _________________ Period: ___ DNA Fingerprinting: Gel Electrophoresis Background Information: Restriction enzymes or restriction endonucleases are essential tools in molelcular biology (specifically for recombinant DNA methodology). Several hundred have been isolated from a variety of prokaryotic organisms. Restriction endonucleases are named according to a specific system of nomenclature. The letters refer to the organism from which the enzyme was isolated. The first letter of the name stands for the genus name of the organism. The next two letters represent the second word or the species name. The fourth letter (if there is one) represents the strain of the organism. Roman numerals indicate whether the particular enzyme was the first isolated, the second, or so on. Examples: EcoRI HaeII E = genus Escherichia co = species coli R = strain RY13 I = first endonuclease isolated H = genus Haemophilus ae= species aegyptus I I = second endonuclease isolated Restriction endonucleases recognize specific DNA sequences in double stranded DNA (usually a four to six base pair sequence of nucleotides) and digest the DNA at these sites. The result is the production of fragments of DNA of DNA of various lengths. Some restriction enzymes cut cleanly through helix at the same position on both strands to produce fragments with blunt ends (figure 6.2a). Other endonucleases cleave each strand off center at specific nucleotides to produce fragments with "overhangs" or sticky ends (figure 6.2b). By using the same restriction enzyme to "cut" DNA from two different organisms, complementary "overhangs" or sticky ends will be produced and allow the DNA from two sources to be "recombined." Figure 6.2a Hae III Cleavage by HaeIII produces blunt ends 5'...GGCC...3' 3'...CCGG...5' Figure 6.2b EcoR I Cleavage by EcoRI produces sticky ends 5'...GAATTC...3' 3'...CTTAAG...5' Modified from: http://www.biologyjunction.com/molecular_biology.htm and Ward’s “DNA Fingerprinting Lab Activity” In this exercise, samples of DNA obtained from a crime scene and two suspects have been incubated with the same restriction enzymes. The resulting fragments of DNA will be separated using gel electrophoresis and compared to a sample of cut to known base pair (bp) lengths. The DNA samples will be loaded into wells of an agarose gel and separated by the process of electrophoresis. After migration of the DNA through an electrical field, the gel will be stained with methylene blue, a dye that binds to DNA. When any molecule enters an electric field, the mobility or speed at which it will move is influenced by the charge of the molecule, the strength of the electrical field, the size and shape of the molecule, and the density of the medium (gel) through which it is migrating. When all molecules are positioned at a uniform starting site on a gel and a gel is placed in a chamber containing a buffer solution and electricity is applied, the molecules will migrate and appear as bands. Nucleic acids, like DNA and RNA, move because of the charged phosphate group in the backbone of the DNA molecule. Because the phosphates are negatively charged at neutral pH, the DNA will migrate through the gel toward the positive electrode. In this exercise, we will use an agarose gel. In agarose, the migration rate of linear fragments of DNA is inversely proportional to their size; the smaller the DNA molecules, the faster it migrates through the gel. General Procedure: A: Preparing the Gel 1. Prepare the agarose gel for electrophoresis according to the directions given by you teacher or in the kit. 2. Obtain the “DNA Marker Std”. The DNA is mixed with a gel-loading solution containing a tracking dye, bromophenol blue, that will make it possible to "track" the processes of its migration in the agarose gel. 3. Obtain the cut DNA from Suspect 1, Suspect 2, and the Crime Scene. The DNA fragments are of a known size will serve as a "standard" for measuring the size of these fragments. It also contains the tracking dye. B: Loading the Gel 1. 2. 3. 4. 5. Helpful Hints for Loading Gel 1. Put a small amount of gel-loading solution into the end of a micropipette. Do not allow the solution to move up into the pipette, or bubbles will be introduced into the well of the agarose gel during loading. 2. Hold the tip of the pipette above the gel and gently dispense the solution. The loading dye is denser than the buffer and will move into the well. (Do not place the tip of the pipette into the well or you might puncture the gel). Pour enough buffer gently over the gel to cover it. Load 10 uL of the DNA Marker Standard (control) into a well. Load 10 uL of the Crime Scene DNA into a second cell. Load 10 uL of Suspect 1’s DNA into a third well. Load 10 uL of Suspect 2’s DNA into fourth well. Modified from: http://www.biologyjunction.com/molecular_biology.htm and Ward’s “DNA Fingerprinting Lab Activity” See the figure below for a side view of a typical gel box. Figure 6.3 Gel Box. C: Electrophoresis: 1. Place the top on the electrophoresis chamber and carefully connect the electrical leads to an approved power supply (black to black and red to red). Set the voltage to the appropriate level for your apparatus. When the current is flowing, you should see bubbles on the electrodes. 2. Allow electrophoresis to proceed until the tracking dye has moved nearly to the end of the gel. 3. After electrophoresis is complete, turn off the power, disconnect the leads, and remove the cover of the electrophoresis chamber. D: Staining and Visualization: Note: Wear Gloves! 1. Carefully remove the gel bed from the chamber and gently transfer the gel to a staining tray for staining. Use the metal spatula under the gel during the transfer. Do not stain in the electrophoresis chamber. 2. Mrs. P will “De-Stain” the gel so that only bands are visible. E: Determining Fragment Size: 1. After observing the gel, carefully wrap it in plastic wrap and smooth out all the wrinkles. 2. Using a marking pen, trace the outlines of the sample wells and the location of the bands with a permanent fine tip marker. 3. Remove the plastic wrap and flatten it out on a white piece of paper on the laboratory bench. Save the gel in a zip lock bag. Add several drops of buffer, store at 4 degrees C. You can make your measurements directly from the marks on the plastic wrap. Modified from: http://www.biologyjunction.com/molecular_biology.htm and Ward’s “DNA Fingerprinting Lab Activity” Analysis and Results Background Information The size of the fragments produced by a specific endonuclease can be determined by using standard fragments of known size. When you plot the data on semilog graph paper, the size of the fragments is expressed in the log of the number of base pairs they contain. This allows data to be plotted on a straight line. The migration distance of the unknown fragments, plotted on the x-axis, will allow their size to be determined on the standard curve. Analyzing Results: 1. Measure the distance of the DNA bands, in millimeters, from the bottom of the sample well to the bottom of each DNA fragment for the DNA marker standard. Measuring to the bottom of each fragment band ensures consistency and accurate measurements. Do not measure the migration distance of the largest fragment nearest the well; it will not be on the standard curve and will skew results. 2. Record the measurements in Table 1 in the Analysis section. 3. On the semi-log graph paper, plot a standard curve for the DNA marker standard. Plot the migration distance in millimeters on the X-axis, against the molecular size in base pairs (bp) of each fragment. Draw the best-fit line to your points. Note: When plotting on semi-log graph paper, the fragment size on the y-axis is expressed on a logarithmic scale. Label the first series of lines 100bp, 200bp, 300bp, etc. Then label the second series of lines as 1000bp, 2000bp, 3000bp, etc. The third series would be 10,000bp, 20,000bp, 30,000bp, etc. And so on. 4. Measure the distance that each band traveled for the lanes containing the two suspects and crime scene DNA. Record the data in the Tables 2, 3, and 4 in the Analysis section. 5. Calculate the base pair size of each of the fragments by moving along the X-axis until you have reached the distance traveled by the fragment. From that point, move upward until you intersect the line of best fit on the graph. Determine where that point is on the Y-axis and estimate the base pair value at that point. Enter this data in the Tables 2-4. Modified from: http://www.biologyjunction.com/molecular_biology.htm and Ward’s “DNA Fingerprinting Lab Activity” Name: __________________________________________ Date: ______________ Period: ____ Analysis: Sketch your results by drawing the bands at the appropriate distances for each. Modified from: http://www.biologyjunction.com/molecular_biology.htm and Ward’s “DNA Fingerprinting Lab Activity” Modified from: http://www.biologyjunction.com/molecular_biology.htm and Ward’s “DNA Fingerprinting Lab Activity” Modified from: http://www.biologyjunction.com/molecular_biology.htm and Ward’s “DNA Fingerprinting Lab Activity” Modified from: http://www.biologyjunction.com/molecular_biology.htm and Ward’s “DNA Fingerprinting Lab Activity” Modified from: http://www.biologyjunction.com/molecular_biology.htm and Ward’s “DNA Fingerprinting Lab Activity” Modified from: http://www.biologyjunction.com/molecular_biology.htm and Ward’s “DNA Fingerprinting Lab Activity” Modified from: http://www.biologyjunction.com/molecular_biology.htm and Ward’s “DNA Fingerprinting Lab Activity”