Insights from community ecology into the role of enemy release in
advertisement

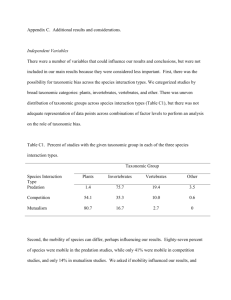
Insights from community ecology into the role of enemy release in causing invasion success: the importance of native enemy effects Biological Invasions Kirsten M. Prior1, Thomas H.Q. Powell, Ashley L. Joseph, Jessica J. Hellmann Corresponding Author1: University of Florida; priorkm@gmail.com Electronic Supplementary Material 1: Additional meta-analysis methods Literature search Systematic literature searches were conducted by searching various combinations of key words in ISI Web of Science, including: “top-down”, “excl*”, “pred*”, “graz*”, “herbivore*”, “parasit*”, “pathogen*”, “marine”, “terrestrial”, “freshwater”. The last comprehensive review of predation effects was performed by Sih et al. (1985), and we also searched for studies that cited this review and other reviews related to top-down control (e.g., Crawley 1989; Lodge 1991; Brett and Goldman 1996; Bigger and Marvier 1999; Halaj and Wise 2001; Shurin et al. 2002). There were no constraints on the journals searched. We only used papers that were published after Sih et al. (1985) and compared our results to this study. After removing studies that did not meet our selection criteria, we were left with a large dataset of 170 studies and 615 “observations” (see below). We compare our results to those of Sih et al. (1985) to help interpret our results. Calculating and compiling effect sizes and statistical methods Effect sizes for each “observation” were calculated using MetaWin 2.1 (Rosenberg et al. 2000). The effect size was calculated as Hedges’ d, the standardized mean difference between prey performance in the absence and presence of native enemies: 𝑑 = [𝑋̅𝐸 − 𝑋̅𝐶 )/s]J, where 𝑋̅𝐸 is the mean of prey performance when enemies are reduced or excluded, 𝑋̅𝐶 is the mean of prey performance in controls when enemies have free access to prey, s is the pooled standard deviation, and J is the small-sample-size bias correction factor (Gurevitch and Hedges 2001). The other commonly used metric in ecological studies, the log response ratio (ln R), was not appropriate for this data set as it contained many zeros and low numbers (Hedges et al. 1999; Salo et al. 2007). The overall results did not differ when conducting analyses with ln R (see Online Resources 4). Effect sizes were calculated for each “observation.” An individual species or a group of species (e.g., chironomids, periphyton) was considered as a single observation in a study. The lowest level of taxonomic resolution was always used. If a study measured the effect of enemies on prey in multiple locations such as in different habitats or at different sites, then each of these locations was retained as an independent observation. This assumes that prey in different locations likely encounter different abiotic and biotic conditions, including different enemy communities (also see Gurevitch et al. 1992). If the experiment manipulated additional factors, such as fertilization levels, we only included treatments under ambient conditions or those closest to ambient conditions. When results were reported in a time series, the final sampling date was used. Peak sampling date was used for some studies of cyclically fluctuating species such as small mammals (as in Salo et al. 2007) or in the case where studies ran until there were low numbers of prey in both controls and exclosures (e.g., for insect populations that peak seasonally). Prey performance was classified as either an effect on biomass or on abundance. In some instances we used total dry weight, percent cover, ash free dry mass, or chlorophyll a as a surrogate for biomass. For responses that represented abundance, variables were: density, abundance, number surviving, or new recruits. We only included studies that examined biomass at the population level, not at the individual level. This enabled us to compare biomass and abundance as measures of the effect of enemies as population-level biomass reflects both the number and the size of a group of individuals. If a study measured biomass and abundance, a composite effect size was calculated by calculating the mean effect size for all observations (Borenstein et al. 2009). Cumulative effect sizes and 95% bias-corrected confidence intervals using resampling tests (4999 iterations) were calculated for the full dataset, each ecosystem, and each taxonomic group within ecosystems (Gurevitch and Hedges 1999). There is an effect of enemy exclusion on prey if the lower confidence interval does not bracket zero. A hierarchical approach was taken to compare effect sizes among groups (as in Gurevitch et al. 1992). Mixed-effects models were run to test the total heterogeneity of the full dataset with no structure, and then to test heterogeneity among ecosystems, and among taxonomic groups within each ecosystem type (Gurevitch and Hedges 2001). Results were compared to a chi-square distribution to determine if the full data set is heterogeneous and if there are significant differences in effect sizes among and within groups. Additionally, to investigate the influence of variation in study design on effect sizes, comparisons were conducted within taxonomic groups among types of response variables (abundance vs. biomass), treatment types (e.g., exclosure vs. insecticide), and level of taxonomic resolution (species vs. community). In addition, we conducted weighted linear regressions between latitude (absolute value), study size (m2), study duration (day) and effect sizes. We found that few effects of study design elements influenced effect sizes (see Online Resources 3). We also conducted comparisons among ecosystems and taxonomic groups at the study level to see if the potential lack of independence of observations within studies influenced our results. We found similar trends in effect sizes when analyzed at the study level (see Online Resources 4). The “file-drawer problem” was assessed using Rosenthal’s Method to calculate fail-safe values for the overall data set and each ecosystem type. These values were compared to Rosenthal’s conservative critical value of 5n + 10, where n is the total number of observations (Rosenberg et al. 2000). Additionally, we created normal quantile plots for the whole dataset and each ecosystem type to assess publication bias and to see if the effect sizes are normally distributed (Wang and Bushman 1998; Rosenberg et al. 2000). Fail-safe numbers were all larger than Rosenthal’s critical value (of the full dataset) of 3085 (full dataset = 73931; terrestrial = 14513; marine = 4450; freshwater = 6590) indicating that the dataset is robust to publication bias. Also, visual assessments of diagnostic plots for the full dataset and each ecosystem type showed few deviations from predicted patterns also suggesting that this data set is robust to publication bias and is normally distributed. Sub-group comparisons Sub-groups were defined based on well-established biological groups or habitat types. For example, taxonomic groups were divided into feeding guilds (e.g., chewing vs. gall-forming vs. sap-sucking insects), growth forms (e.g., crustose vs. macroalgae), and/or taxonomic groups (e.g., mollusk vs. crustacean vs. annelid marine invertebrates). Invertebrate, vertebrate and pathogen (for plants only) enemies were compared for each taxonomic group. Enemy groups were further divided into feeding guilds (e.g., specialist vs. generalist invertebrates), and/or taxonomic groups (e.g., bird vs. reptile insectivores). Finally, within ecosystem type, observations were categorized by climate region (e.g., temperate vs. tropical) and by habitat type (e.g., forest vs. grassland; reef vs. intertidal; lentic vs. lotic). Observations were assigned to subgroups based on available information in studies or by referring to external sources such as the USDA Plants Database. The goal for this sub-group analysis was to search for patterns that could further explain variation in native effects within major taxonomic groups. Effect sizes and analyses were conducted as described above. A conservative P value (P = 0.01) was used to account for making multiple comparisons on the same dataset (Gates 2002). A hierarchical approach was taken in some instances. For example, for terrestrial plant sub-groups, we compared graminoids, forbs and woody plants. We then sub-divided these groups further, comparing annual and perennial graminoids, and annual and perennial forbs. Groups were only included in analyses if the contained five or more observations (Gates 2002). Sub-groups were not always mutually exclusive; however, making comparisons among multiple sub-groups allowed us to search for prominent patterns in the dataset. In all but one case there were either not enough studies, or not enough groups to conduct sub-group comparisons within vertebrate prey groups. References Borenstein M, Hedges LV, Higgins JPT, Rothstein HR (2009) Introduction to Meta-analysis. John Wiley and Sons, Chichester Bigger DS, Marvier MA (1999) How different would a world without herbivory be?: A search for generality in ecology. Integr Biol 1:60-67 Brett MT, Goldman CR (1996) A meta-analysis of the freshwater trophic cascade. Proc Natl Acad Sci USA 93:7723-7726 Crawley, M. (1989) Insect herbivores and plant population dynamics. Annu Rev Entomol 34:531–564 Gates, S. (2002) Review of methodology of quantitative reviews using meta-analysis in ecology. J Anim Ecol:71:547-557. Gurevtich J, Hedges LV (2001) Meta-analysis: combining the results of independent experiments. In: Scheiner S, Gurevitch J (eds) Design and analysis of ecological experiments. Chapman and Hall, New York, pp 347-369 Gurevitch J, Morrow LL, Wallace A, Walsh JS (1992) A meta-analysis of competition in field experiments. Am Nat 140:539 Halaj J, Wise DH (2001) Terrestrial trophic cascades: how much do they trickle? Am Nat 157:262-281 Hedges LV, Gurevitch J, Curtis PS (1999) The meta-analysis of response ratios in experimental ecology. Ecology 80:1150-1156 Lodge DM (1991) Herbivory on freshwater macrophytes. Aquat Bot 41:195-224 Rosenberg MS, Adams DC, Gurevitch J (2000) MetaWin: Statistical Software for Meta-analysis, Version 2.0. Sinauer Associates, Massachusetts Salo P, Korpimäki E, Banks PB, Nordström M, Dickman CR (2007) Alien predators are more dangerous than native predators to prey populations. Proc R Soc B 274:1237-1243 Sih A (1985) Predation, competition, and prey communities: a review of field experiments. Annu Rev Ecol Syst 16, 269–311 Shurin JB, Borer ET, Seabloom EW, Anderson K, Blanchette CA, Broitman B et al (2002) A cross-ecosystem comparison of the strength of trophic cascades. Ecol Lett 5:785–791 Wang MC, Bushman BJ (1998) Using normal quantile plot to explore meta-analytic data sets. Psychol Methods 1:46-54