tshvbh_myvl_ltrktnbrg
advertisement
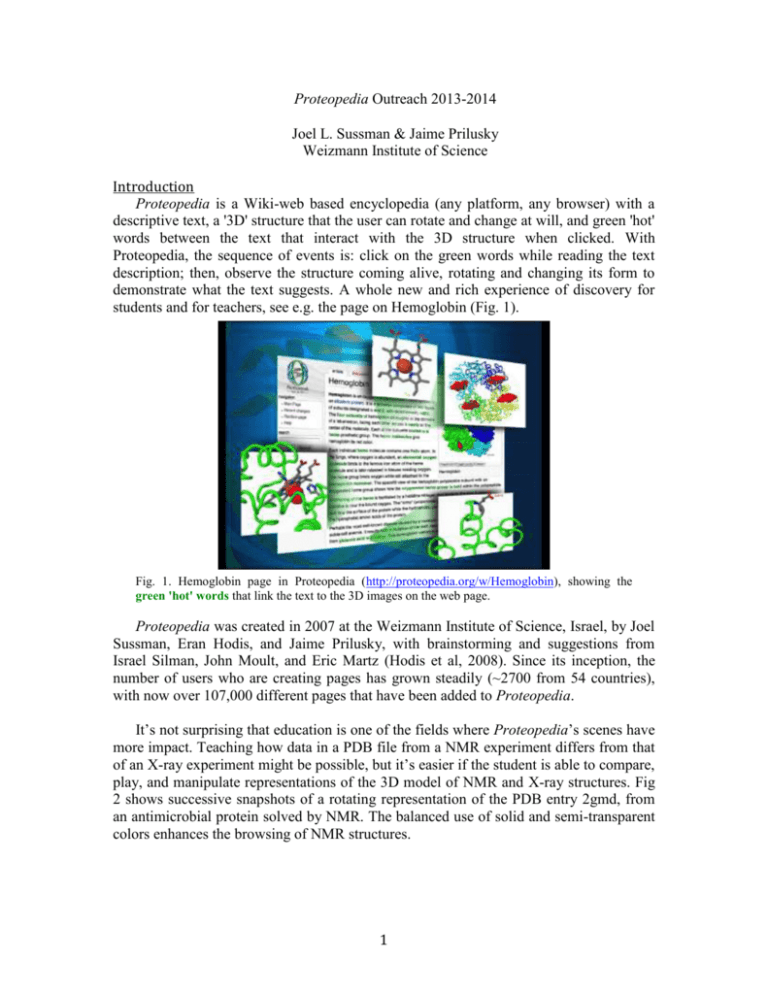
Proteopedia Outreach 2013-2014 Joel L. Sussman & Jaime Prilusky Weizmann Institute of Science Introduction Proteopedia is a Wiki-web based encyclopedia (any platform, any browser) with a descriptive text, a '3D' structure that the user can rotate and change at will, and green 'hot' words between the text that interact with the 3D structure when clicked. With Proteopedia, the sequence of events is: click on the green words while reading the text description; then, observe the structure coming alive, rotating and changing its form to demonstrate what the text suggests. A whole new and rich experience of discovery for students and for teachers, see e.g. the page on Hemoglobin (Fig. 1). Fig. 1. Hemoglobin page in Proteopedia (http://proteopedia.org/w/Hemoglobin), showing the green 'hot' words that link the text to the 3D images on the web page. Proteopedia was created in 2007 at the Weizmann Institute of Science, Israel, by Joel Sussman, Eran Hodis, and Jaime Prilusky, with brainstorming and suggestions from Israel Silman, John Moult, and Eric Martz (Hodis et al, 2008). Since its inception, the number of users who are creating pages has grown steadily (~2700 from 54 countries), with now over 107,000 different pages that have been added to Proteopedia. It’s not surprising that education is one of the fields where Proteopedia’s scenes have more impact. Teaching how data in a PDB file from a NMR experiment differs from that of an X-ray experiment might be possible, but it’s easier if the student is able to compare, play, and manipulate representations of the 3D model of NMR and X-ray structures. Fig 2 shows successive snapshots of a rotating representation of the PDB entry 2gmd, from an antimicrobial protein solved by NMR. The balanced use of solid and semi-transparent colors enhances the browsing of NMR structures. 1 Fig. 2. View of PDB entry 2gmd (http://proteopedia.org/w/2gmd) (Japelj et al, 2007) Proteopedia. Outreach to high school & Undergraduates A series of workshop and have been held at the Davidson Institute and the Science Teaching Center at the Weizmann Institute of Science. These include one for High School Student in April 2014 conducted by Dr. Yael Schwartz of the Science Teaching Center (Fig. 3) and another by Dr. Angel Herraez (from Dept. of Systems Biology, University of Alcalá, Spain) for the members of the of the Science Teaching Center. Fig. 3. High-School students from throughout Israel learning to use Proteopedia at the workshop held in the Davidson Center on 6-Apr-2014. High-school students have also come on a regular basis to the Weizmann Institute on for special tutorials on Proteopedia and working with physical models of proteins, e.g. 7th Graders from the Gifted Teen Center in Emek Hefer in May 2014 (Fig. 4) (see also (Appendix – I) of warm letter of thanks from the students parents). 2 (a) (b) (c) th Fig. 4. 7 Graders from Gifted Teen Center in Emek Hefer visiting the Weizmann Institute of Science (23-May-2014) (a) Joel Sussman with two students and their teacher, Irit Levi examining physical models of 3D protein structures; (b) Students learning Proteopedia; (c) Shai Biran (PhD student at the Weizmann Institute) giving a short lecture in the 3D Molecular Visualization Theatre at the Weizmann Institute to the 7th Grade Students and their teacher. Several of the Proteopedia pages have been translated into Hebrew and Arabic: List of Hebrew pages: http://proteopedia.org/wiki/index.php/Entry_list_(Hebrew) Pages in Arabic: o http://www.proteopedia.org/w/1eve (Arabic) o http://proteopedia.org/w/Beta_Secretase (Arabic) o http://www.proteopedia.org/w/Lysozyme (Arabic) o http://www.proteopedia.org/w/Myoglobin (Arabic) In 2013, Jaime Prilusky and Joel Sussman were guest editors to a special issue of the Israel Journal of Chemistry in which they also published an article on “JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia” (Hanson et al, 2013). Proteopedia as well as ConSurf (from Nir Ben-Tal’s lab) were featured on the cover of that issue of the Journal (Fig. 5). 3 Fig. 5. The cover of The Israel Journal of Chemistry (Vol 53, Issue 3-4) shows on the left a ConSurf analysis of the influenza neuraminidase protein. The 3D tetramer colored by conservation grades is shown in (A) with maroon indicating the most conserved amino acids, and close-up views (B, C) are shown of one of the conserved regions with the anti-flu drug, oseltamivir (tamiflu) bound (Celniker et al, 2013); On the right is a page from Proteopedia as viewed on an iPad, via the JSmol viewer, showing a complex of HIV-Protease and Saquinavir (Invirase), the first protease inhibitor approved by the FDA for the treatment of HIV (Hanson et al, 2013). During the Fall Semester 2014-15, Jaime Prilusky and Joel Sussman will be giving a one-semester course at the Hebrew University of JerusalemFaculty of Agriculture (Rehovot Branch) to undergraduates and MSc student entitled: “Proteopedia – in silico lab course”. The syllabus of the course, in Hebrew, is attached (Appendix – II) Jaime Prilusky & Joel Sussman also gave a 2-day workshop on Proteopedia at the Biennial Conference on Chemical Education at the Grad Valley State University, Allendale, MI on (http://bcce-submissions.com/showSympPap.php?id=773) in August, 2014. 4 References: Celniker G, Nimrod G, Ashkenazy H, Glaser F, Martz E, Mayrose I, Pupko T, Ben-Tal N (2013) ConSurf: Using Evolutionary Data to Raise Testable Hypotheses about Protein Function. Israel J Chem 53: 199-206 Hanson RM, Prilusky J, Renjian Z, Nakane T, Sussman JL (2013) JSmol and the NextGeneration Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Israel J Chem 53: 207-216 Hodis E, Prilusky J, Martz E, Silman I, Moult J, Sussman JL (2008) Proteopedia – a scientific ‘wiki’ bridging the rift between 3D structure and function of biomacromolecules. Genome Biol 9: R121 Japelj B, Zorko M, Majerle A, Pristovšek P, Sanchez-Gomez S, de Tejada GM, Moriyon I, Blondelle SE, Brandenburg K, Andrä J, Lohner K, Jerala R (2007) The Acyl Group as the Central Element of the Structural Organization of Antimicrobial Lipopeptide. J Am Chem Soc 129: 1022-1023 5 Appendix I –Parents’ letter of 7th Graders from the Gifted Teen Center in Emek Hefer 6 Appendix II –Syllabus for Proteopedia – in silico lab course at the Faculty of Agriculture of the Hebrew University in Jerusalem (Rehovot Branch) 7 8