emi12305-sup-0001-si
advertisement

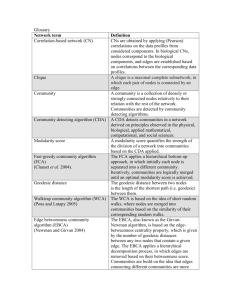
Table S1. Brief explanation of network analysis terms used in the manuscript1 Term Assortativity coefficient Explanation Tendency of the nodes to attach to nodes with similar degrees. A negative value indicates a tendency of nodes to attach to nodes with different degrees. Cellular automata Mathematical model consisting of a regular grid of cells. Each cell has a defined state at a particular time, which is updated periodically according to the states of the neighbouring cells. Cellular automata are very useful to model the competition for space of different strains in time. Connectivity mechanisms The mechanisms by which nodes connect to form a network. Connection Also called an edge. An interaction between two nodes. It can have a direction, if a node acts towards another, or it can be undirected if both nodes act equally in the interaction. In the antagonistic interaction network presented in this article, all connections are directed. Degree Total number of connections a node has to other nodes in the network. It is the sum of: the in-degree (receiver degree), or the number of connections coming to a node and the out-degree (sender degree), or the number of connections stemming from a node. Degree distribution Distribution of the degrees of all nodes in the network. Directed connection A connection going from one node to another one. Also called directed edge. Example: A strain antagonizes another one. Disassortative network A network where nodes attach preferentially to nodes with different degrees. Disassortative networks have negative assortativity coefficients Dyad A sub-network made of two nodes. Edge A connection between two entities (or nodes) in a network. For example, an antagonistic interaction. Erdős and Rényi networks (ER networks) Random networks commonly used as null models in network analysis. Hyperimmunity A state where many strains have antagonistic mechanisms, but most strains are resistant to them. Interaction density Fraction of strains with observed interactions relative to the number of possible strain-pairs Nestedness Tendency of the nodes to attach with subsets of the connections of more connected nodes. In a highly nested network, nodes with few connections tend to connect with nodes with many connections, rather than with nodes with intermediate numbers of connections. Network diameter Largest number of antagonistic interactions needed to connect any two strains by the shortest path between them. To find the diameter one can count the connections needed to connect all pairs, and take the maximum. For example, in the three member network: A->B->C, to connect A with C one has to go from A to B and from B to C. Thus, two connections are needed to go from A to C, and the network diameter is 2. Network size Number of nodes of a network. Node An entity in a network, (in our case a strain). They are also called vertexes. Nodes are connected through edges. Non-transitive antagonistic interaction loops Given a directed interaction from one member towards another one, and the latter with a third one, the interaction is non-transitive if it is not present from the first towards the third one, or if it is present from the third towards the first one. Transitive: A -> B and B -> C implies A -> C Non-transitive: A -> B and B -> C but A C Non-transitive: A -> B and B -> C but A <- C Table S1. Continuation Preferential attachment A mechanism of network growth, where new nodes connect to already highly connected nodes with higher probability than to less connected nodes. The result is that already more connected nodes become even more connected. Poisson degree distribution A distribution that is often used to model the number of events that occur in a fixed time interval when the events occur at random but with fixed rate. In the case of network analysis, instead of events, connections are modelled that are formed between pairs of a fixed number of nodes, with a fixed probability of formation of each node (or a fixed number of total nodes). Power-law distribution A distribution of a random variable whose probability density function is characterized by a power of the variable. These distributions have slowly-decaying tails, which means that large values of the random variable are observed with non-negligible probabilities. Receiver degree See Degree Scale-free (SF) network SF networks are characterized by having power-law degree distributions in their right tails. This implies that they have a few nodes with very large degrees, while most nodes have only small degrees. Sender degree See Degree Sender determined network See Sender-receiver asymmetry Sender-receiver asymmetry Comparison of the variance of the antagonism and sensitivity degrees to determine weather an interaction network is on average determined more by the potential of inhibition or by the sensitivity of strains. Negative values indicate a sender determined interaction network. Small world network A type of network where the diameter is small in comparison with the number of nodes, so that all the nodes can be connected by a small number of edges. In small world networks, the diameter varies logarithmically with network size. Sub-network A subset of the nodes of a network and the connections between them. Tail of a distribution Behaviour of a distribution at the extremes. In network analysis, the right tail, on the higher values, is usually referred to when not indicated otherwise. Triad A sub-network made of three nodes. 1. These explanations are intended to help the reader understand the technical terms related to network analysis used in the text, and are not formal or mathematical definitions. Table S2. Abbreviated vs. full strain names Abbreviated name 1A6 1B6 1B7 1B8 1C11 1C9 1D12 1D4 1D6 1D8 1D9 1E10 1E2 1E3 1E6 1E7 1E8 2A1 2A4 2B1 2B4 2B7 2C1 2C2 2C5 2C8 2C9 2D8 2D9 2E4 3E2 3E3 4A4 4B2 4B3 4B8 4D1 Full strain name J5.1A6 J4.1B6 J4.1B7 J4.1B8 J3.1C11 J3.1C9 J2.1D12 J2.1D4 J2.1D6 J2.1D8 J2.1D9 J1.1E10 J1.1E2 J1.1E3 J1.1E6 J1.1E7 J1.1E8 J5.2A1 J5.2A4 J4.2B1 J4.2B4 J4.2B7 J3.2C1 J3.2C2 J3.2C5 J3.2C8 J3.2C9 J2.2D8 J2.2D9 J1.2E4 J1.3E2 J1.3E3 J5.4A4 J4.4B2 J4.4B3 J4.4B8 J2.4D1 Figure S1. Nestedness analysis matrix. Rows represent antagonist strains and columns represent sensitive strains. Rows and columns were ordered with the algorithm of Rodríguez-Gironés & Santamaría (2006) to maximize nestedness visually. Black squares represent antagonistic interactions from the corresponding rows to the corresponding columns. Figure S2. Network diameter as a function of network size in simulated networks using our model (Equations 1 and 2). Values (black dots) are means of 100 simulations. The grey line indicates a logarithmic fit (y = 0.551log(x) + 1.3182). Logarithmic increase of the network diameter with network size is a hallmark of small-world type networks (Amaral et al., 2000). Figure S3. Parameter space analysis of observed and simulated networks. Shades represent triad counts with different parameter combinations, for the sixteen possible triads (A to P), according to the corresponding scale above each panel. For parameter space resolution and simulation numbers see Methods. Red circles are located in the axes according to the values observed in the actual network. Shades inside these circles indicate the observed triad counts. 1 2 4 3 5 Figure S4. Example of the scale used. Values below the images show the rating assigned to each image.