Paper - Lawrence Lab
advertisement

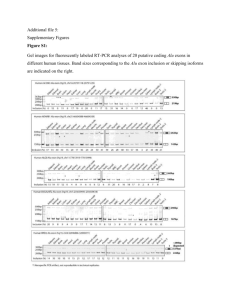
Myhal 1 Alexandra Myhal, Eric Ottesen, and Dr. Ravindra Singh Iowa State University Biomedical Science Division George Washington Carver Internship Program 2011 Research Title Quantification of Human SMN Spliced Variants by qPCR Abstract Quantitative PCR (qPCR) is an accurate method of determination of relative abundance of transcripts expressed in normal and pathological conditions in a particular cell type. Here I used the Stratagene MxPro 3005P qPCR machine from Agilent Technology to determine the level of several spliced isoforms of the survival motor neuron (SMN) gene. We particularly focused on inclusion and skipping of exon 7, which is associated with spinal muscular atrophy (SMA), a leading genetic disease of children and infants. The SMN gene is present in two copies, SMN1 and SMN2, which are differentially spliced. SMA occurs when SMN2 is the only gene present, as it is unable to compensate for the loss of SMN1 due to increased skipping of exon 7 (Ref. 2). Our findings underscore that exon 5 and exon 7 are more susceptible to skipping under oxidative stress. Further study will be needed to determine the extent of skipping of different exons in different tissues. Introduction The goal of this paper is to quantify human SMN spliced variants by qPCR. There are two copies of the SMN gene, SMN1 and SMN2, with only one nucleotide difference Myhal 2 between them. When SMN2 undergoes splicing there is an increase in exclusion of exon 7 and as a result SMA occurs when SMN2 is the only copy present. A healthy human has both copies of the SMN gene while a SMA patient only has SMN2. The objective of this research is to quantify SMN spliced variants into two different treatments of neuronal SHSY-5Y cells. To quantify the spliced variants we used qPCR, which measures the total amount of PCR product after each cycle of amplification. Two qPCR experiments were completed, the Relative Quantity and Standard Curve Quantification. The Relative Quantity assay measures the relative concentration of the isoform of interest. The Standard Curve Quantification assay provides absolute quantification of each template. With the resulting data from both assays we were able to conclude that there was an increased amount of skipping of exon 5 and exon 7 in neuronal SHSY-5Y cells in conditions of oxidative stress. Results Different assays were conducted to compare the difference isoforms between normal neuronal SHSY-5Y cells and neuronal SHSY-5Y cells treated with paraquat, which causes conditions of oxidative stress. A. Paraquat - + Myhal 3 First, we identified the isoforms of interest (FL, ∆7, ∆5, and ∆57) by Reverse Transcription- PCR (RT-PCR) from exon 4 to 8 (figure A.). The relative intensity of each isoform was identified and we then started qPCR assays for each isoform. B The first assay completed was the Relative Quantity assay. This assay measures the concentration of the SMN spliced variants. The calibrator is the first bar in each assay, which is the baseline for the expression of a certain isoform. The untreated neuronal SHSY-5Y cells were the calibrator. To normalize the assay we used GAPDH gene. The results are “…the fold difference between the targets and normalizer in experimental versus calibrator samples” (1). This chart shows that there was an increase in relative quantity of isoform ∆5, ∆7, and ∆57 in neuronal SHSY-5Y cells treated with paraquat. There is a decrease in relative quantity of full length SMN isoform in neuronal SHSY-5Y cells treated with paraquat. Above each assay is the region of interest and the exon-exon junctions where the primers annealed. Myhal 4 100.00 Relative Abundance C 10.00 1.00 0.10 Untreated Treated FL avg 40.14 28.85 ∆7 avg 12.85 16.51 ∆5 avg 1.02 1.63 ∆57 avg 0.57 1.61 The Standard Curve Quantification test provides the template quantity for each template. The absolute quantity of each isoform was determined against a serial dilution of plasmid DNA. The relative abundance was determined by taking the percentage of each isoform compared to the total expression of SMN (SMN Universal). Each bar represents the average of the three replicate reactions. This logarithmic graph shows there is increased abundance of the ∆7, ∆5, and ∆57 isoforms in the paraquat treated neuronal SHSY-5Y cells. There is slso a decrease in the FL isoform in the paraquat treated neuronal SHSY-5Y cells. Materials and Methods Myhal 5 The neuronal SHSY-5Y cells were exposed to one 1mM of paraquat for 24 hours which induced conditions of oxidative stress. RNA was isolated from paraquat and PBS treated cells by Trizol (Invitrogen) and the cDNA was generated using SuperScript (Invitrogen) III Reverse Transcriptase using the manufacturer’s protocol. The qPCR machine used for this research was the Stratagene MxPro 3005P and the data was analyzed by MxPro QPCR Software version 4.10 both from Agilent Technology. The Master mix for each assay reaction used was the Brilliant II SYBR Green QPCR Master mix manufactured by Agilent Technologies. The primer sets for each assay were designed to anneal to specific exon-exon junctions to distinguish different spliced isoforms. Each cDNA sample was amplified in triplicate for every assay. The control group for each absolute quantification assay was a standard curve produced by six to eight 4-fold serial dilutions of plasmid. Plasmids specific for each isoform were generated by amplifying SMN Exons 2b8, then the bands corresponding to each isoform were gel purified and ligated into the pGEM-T Easy vector (Promega). The control plasmd for the SMN Universal assay was generated by amplifying cDNA using the 5’ Exon 1 and 3’ Exon 1+2a primers and ligated into pGEM-T Easy. The following isoforms were amplified using these primers: Isoform FL: 5’ Exon 5+6- TTC CTT CTG GAC CAC CAA TAA and 3’ Ex 7- CAC CTT CCT TCT TTT TGA TTT TGT C, Isoform ∆7: 5’ Exon 5+6- TTC CTT CTG GAC CAC CAA TAA and 3’ Exon 6+8- TGC TCT ATG CCA GCA TTT CCA TAT, Isoform ∆5: 5’ Exon 4+6AGG TGG TGG GGG AAT TAT CTT T and 3’ Ex 7- CAC CTT CCT TCT TTT TGA TTT TGT C, Isoform ∆57: Exon 4+6- AGG TGG TGG GGG AAT TAT CTT T and 3’ Myhal 6 Exon 6+8- TGC TCT ATG CCA GCA TTT CCA TAT, and Isoform SMN U: 5’ Exon 1-TGC TAT GGC GAT GAG CAG C and 3’ Exon 1+2a-CAG AAT CAT CGC TCT GGC CTG T. The splice variants targeted are ∆ 7, ∆ 5, ∆ 57, Full Length, and SMN universal. As normalizing assay, GAPDH was amplified using the following primers: hGAPDH-1-AAC AGC GAC ACC CAC TCC TC and hGAPDH-2-CAT ACC AGG AAA TGA GCT TGA CAA (Figure B). Discussion Our results confirm an inhibitory effect of paraquat on SMN splicing. We found a higher co-skipping of exon 5 and 7 skipping of either exon 5 or 7 in the neuronal SHSY5Y cells under oxidative stress. Our results suggest a possible correlation between skipping of exons 5 and 7. These findings indicate that SMA patients exposed to paraquat would have increased severity of disease. QPCR was used instead of RT-PCR in this research because it provides more concrete information about the spliced variants and the results are numeric. The absolute quantification of each spliced variant provided more information about the SMN gene. The results from this research will give scientists a better understanding of SMN gene splicing products and will allow further their work towards a cure for SMA. Conclusion Myhal 7 Our quantitative assays showed increased skipping of exon 5 and exon 7 in the neuronal SHSY-5Y cells under conditions of oxidative stress. Also, there is a decrease in skipping of the Full Length Isoform. References 1. Stratagene (2007). Introduction to Quantitative PCR. Methods and Applications Guide. 2. Monani, U. R., Lorson, C. L., Parsons, D. W., Prior, T. W., Androphy, E. J., Burghes, A. H. M., & McPherson, J. D. (1999). A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from copy gene SMN2. Human Molecular Genetics, Volume 8 (issue 7), 1177-1183.