Phylogenetic Trees
advertisement
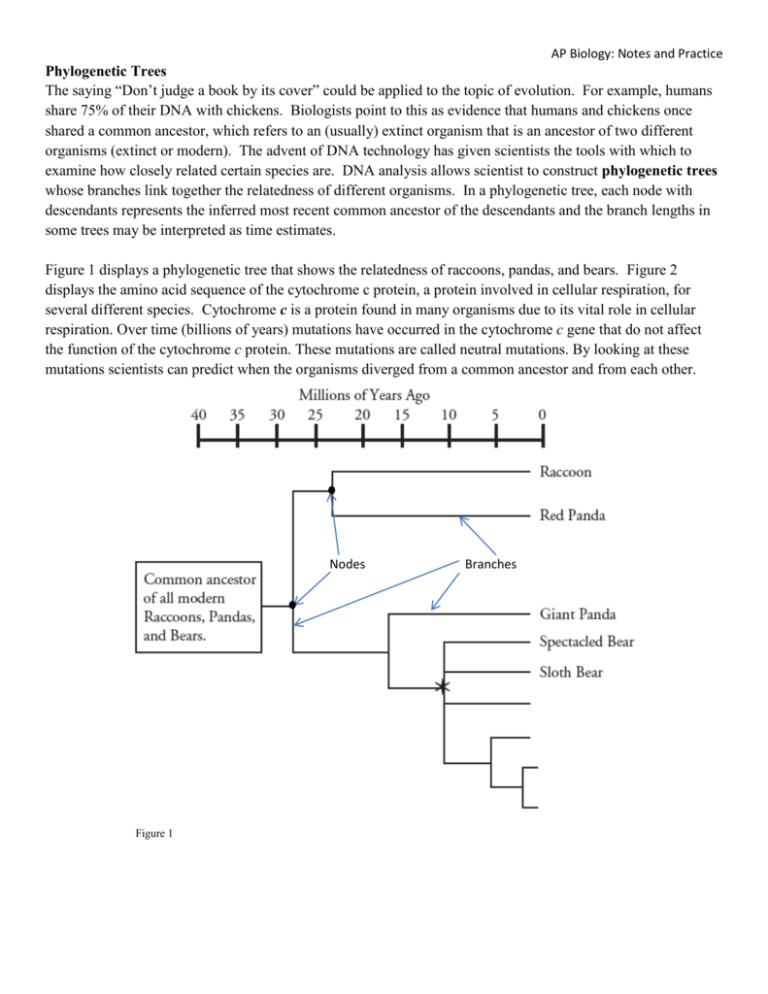
AP Biology: Notes and Practice Phylogenetic Trees The saying “Don’t judge a book by its cover” could be applied to the topic of evolution. For example, humans share 75% of their DNA with chickens. Biologists point to this as evidence that humans and chickens once shared a common ancestor, which refers to an (usually) extinct organism that is an ancestor of two different organisms (extinct or modern). The advent of DNA technology has given scientists the tools with which to examine how closely related certain species are. DNA analysis allows scientist to construct phylogenetic trees whose branches link together the relatedness of different organisms. In a phylogenetic tree, each node with descendants represents the inferred most recent common ancestor of the descendants and the branch lengths in some trees may be interpreted as time estimates. Figure 1 displays a phylogenetic tree that shows the relatedness of raccoons, pandas, and bears. Figure 2 displays the amino acid sequence of the cytochrome c protein, a protein involved in cellular respiration, for several different species. Cytochrome c is a protein found in many organisms due to its vital role in cellular respiration. Over time (billions of years) mutations have occurred in the cytochrome c gene that do not affect the function of the cytochrome c protein. These mutations are called neutral mutations. By looking at these mutations scientists can predict when the organisms diverged from a common ancestor and from each other. Nodes Figure 1 Branches AP Biology: Notes and Practice Figure 2 1. Describe what the nodes and branches in Figure 1 represent. 2. According to Figure 1: a. How long ago did the common ancestor of all the organisms on this phylogenetic tree exist? b. How long ago did giant pandas and spectacled bears diverge from a common ancestor? AP Biology: Notes and Practice c. What animal shares the most recent common ancestor with the red panda? Explain your reasoning. 3. Which are more closely related, the Giant Panda and the Red Panda or the Giant Panda and the Sloth Bear? Justify your answer. 4. According to the passage, how did scientists likely determine the placement of each organism on the phylogenetic tree in Figure 1? 5. In addition to determining the relatedness of species by comparing DNA sequences, phylogenetic trees can be constructed using morphological (structural) characteristics. Which of the following pieces of morphological data would have been the least useful for a scientist in constructing this phylogenetic tree? Explain your answer. A. Pandas have wrist bones that extend further than bear wrist bones B. Pandas, red pandas, and raccoons have tooth patterns different from other bears C. Raccoons, pandas, and bears all contain a radius and ulna (bones of the forearm) that are separate, allowing the animals to rotate their limbs D. Red pandas and raccoons share a similar skull pattern, especially in terms of jaw bone development 6. The Sloth Bear was found to have 5 specific morphological traits. Four additional bears (the Sun Bear, Brown Bear, Polar Bear, and Black Bear) were examined for these same traits. The presence (+) or absence (-) of each trait is displayed in the table below: Trait Sloth Bear Sun Bear Brown Bear Polar Bear Black Bear 1 + + + 2 + + - 3 + + - 4 + + + + + 5 + + + + + The phylogenetic tree in Figure 1 contains 4 unlabeled branches. Using the information in the table, place the additional bears in the appropriate unlabeled branch on the phylogenetic tree. Explain your reasoning below. AP Biology: Notes and Practice 7. According to Figure 2: a. How many different species are represented in the cytochrome c amino acid sequences? b. What do the individual letters represent? c. What do the asterisks represent? 8. According to the passage, why do all of the organisms listed in Figure 2 produce cytochrome c? 9. The passage states that DNA analysis allows scientists to construct phylogenetic trees, but Figure 2 provides data from a protein analysis. Can the data in Figure 2 be used to construct a phylogenetic tree? Why or why not? 10. Circle the pairing of organisms from Figure 2 that you predict would have the most divergence in their DNA. Support your choices with a short statement about morphology (observable characteristics and structure), ecology (habitat), or lifestyles of the animals. a. whale and human or whale and tuna b. pig and rhesus monkey or pig and chicken c. tuna and fly or tuna and chicken 11. In Figure 2, the amino acid positions shaded in gray indicate that all 7 organisms have the same amino acid in this position. Write down the number of differences between the amino acid sequence of cytochrome c in a human and each of the following organisms. Organism # of amino acid diff. Tuna Whale Rhesus Monkey Chicken Pig Yeast Fly AP Biology: Notes and Practice 12. The fewer the number of amino acid sequence differences between two organisms, the more recently these two organisms are likely to have diverged from a common ancestor. Based on the differences in amino acid sequences identified in question 11, to which organism from the list are humans: a. Most closely related to? b. Most distantly related to? 13. Enter the names of the seven organisms in Figure 2 into the tree below, to illustrate a phylogenetic relationship between these organisms and humans, as suggested by the cytochrome c data. 14. How do your predictions about relatedness based on morphology, ecology, and lifestyle in Question 10 compare to the relatedness information in the phylogenetic tree developed in Question 13? What conclusion can be made regarding the dependability of morphological, ecological, and lifestyle evidence on determining the relatedness of species? 15. A scientist argues that evaluation of amino acids 68-73 is more reliable than evaluation of amino acids 7883. Do you agree or disagree with this statement? Explain your answer. AP Biology: Notes and Practice 16. DNA evidence suggests that the sequence for beta-casein, a protein found in milk, is highly conserved among mammals. The table below displays the number of sequence differences between 6 different mammals. Construct a phylogenetic tree between these organisms to reflect relatedness as indicated by the beta-casein protein data. Sperm Whale Porpoise Hippopotamus Cow Pig Deer Sperm Whale 0 Porpoise 2 0 Hippopotamus 3 3 0 Cow 8 8 8 0 Pig 10 10 11 13 0 Deer 10 10 10 4 13 0