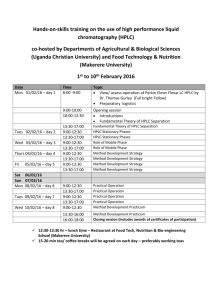
tpj12570-sup-0003-Supp-Legends
advertisement

Supporting information legends Ariizumi et al. Figure S1. Comparison of seedling growth and fruit coloration between WT and pyp1 mutants. (a) Plant growth in the WT and pyp1 mutants. No clear difference in growth was found between 40-d-old WT and pyp1 mutant plants. Bar = 5 cm. (b) Time course of fruit development in the WT and pyp1 mutants. Fruits at different days after flowering (DAF) were compared between the WT and two pyp1 mutants. No significant difference in fruit coloration was found. Bar = 2 cm. Figure S2. Sequence alignment of PYP1 and its homologues. Alignment among the amino acid sequences of PYP1 (Solyc01g098110), its tomato homologue (Solyc02g094430), Arabidopsis PES1 (At1g54570), and PES2 (At3g26840) was performed using Genetyx-MAC Multiple-Alignment (http://www.genetyx.co.jp/). N-terminal regions were relatively less conserved among these proteins. Figure S3. Phylogenetic analysis of PYP1 orthologues in plants. Full-length amino acid sequences of PYP1 and 17 orthologous proteins from 15 plants species were phylogenetically analyzed and evolutionarily related proteins were estimated using Neighbor Joining (NJ) phylogenetic (http://www.phylogeny.fr/version2_cgi/one_task.cgi?task_type=bionj). One analysis hundred bootstrap replicates were performed. Tomato PYP1 is highlighted in red, while plants that bear yellow petals are indicated in yellow. Functionally characterized Arabidopsis PES proteins are marked with an arrow. Bar indicates estimated evolutionary distance, calculated by amino acid substitutions per site. Accession numbers of protein sequences shown are given in Table S8. Figure S4. Expression analysis of PYP1. (a) Semi-quantitative RT-PCR analysis of PYP1. Expression of SAND was used as a loading control. n, negative control. (b) qRT-PCR analysis of PYP1 mRNA expression levels in the WT and pyp1 mutants. The analysis was performed using real-time PCR. Statistical analysis was performed using ANOVA (p < 0.05). Error bars indicate standard error (SE, n=3). PYP1 mRNA levels were significantly different at stages 2 and 3 in pyp1-1 compared to the WT and pyp1-2. Figure S5. Identification of carotenoid fractions in mature petals of the WT and pyp1-1 mutant by HPLC analysis. (a) HPLC chromatographs of non-saponified petal samples from the WT (above) and pyp1-1 mutant (below). (b) HPLC chromatographs of saponified petal samples from the WT (left) and pyp1-1 mutant (right). The numbers above peaks indicate carotenoids identified by HPLC. These numbers correspond to peak numbers (PNs) as in Figure 4. Y-axis indicates milli-absorbance units (mAU). Figure S6. Profiles of carotenoids and their derivative esters present in WT and pyp1 anthers. Non-saponified (left column) and saponified (right column) carotenoid extracts from mature anthers were analyzed by HPLC. Carotenoid esters were detected in saponified WT anthers, while no such esters were detected in the pyp1 mutant anthers. The peaks indicated by asterisks correspond to β-carotene, which is not classified as a carotenoid ester. The numbers above peaks indicate carotenoids identified by HPLC. These numbers correspond to peak numbers (PNs) as in Figure 4. Y-axis indicates micro-absorbance units (mAU). Figure S7. Comparison of contents of each carotenoid in petals at stage 2 between the WT and pyp1 mutant. (a) Quantitative analysis of carotenoids and their ester derivatives at stage 2 in samples not subjected to saponification. (b) Quantitative analysis of carotenoids at stage 2 in samples subjected to saponification. The peak numbers (PNs) listed on the x-axis indicate each carotenoid compound detected by HPLC analysis, and correspond to those in Figure 4 or Figure S4. Error bars indicate standard error (SE, n=3). Mean comparison tests (Student’s t test, P < 0.05) were performed. Asterisks indicate significantly difference between the WT and pyp1 mutant. Figure S8. Comparison of mRNA expression of carotenogenic genes between the WT and two pyp1 mutant alleles. The cDNAs derived from three stages as in Figure 1 were used in the qRT-PCR analysis. S1, stage 1; S2, stage 2; S3, stage 3. Error bars indicate standard error (SE, n=3). All experiments were performed in biological triplicate for each time point and plant. Statistical analysis was performed using ANOVA (P < 0.05), although no significant differences in mRNA expression levels were found between the WT and two pyp mutants at any time point examined. Figure S9. Complementation experiment verifying that Solyc01g098110 is the gene responsible for the pyp1 mutant. (a) Southern blot analysis of sixteen independent complementation lines (pyp1 mutant transformed with full-length PYP1 cDNA). (b) Restoration of flower color in three complementation lines (line A, K, O). Bar = 1 cm. (c) HPLC chromatograph of WT (above) and line O (below) by saponification (right) and by non-saponification (left). The peaks indicated by asterisks correspond to β-carotene, which is not classified as a carotenoid ester. Y-axis indicates milli-absorbance units (mAU). (d) Petal chromoplast morphology in line O at stage 3. Bar = 200 nm. Table S1. Genetic analysis indicates that the pyp1 mutations are monogenic recessive. F2 progeny were obtained from F1 plants of crosses between pyp1 mutant and either Micro-Tom or Ailsa Craig and were sown for phenotypic examination. Both Micro-Tom and Ailsa Craig are WT plants with no mutations in PYP1. Table S2. Primer sequences used for this study. DXS, 1-DEOXY-D-XYLULOSE 5-PHOSPATE SYNTHASE; GGPS2, CHROMOPLAST- SPECIFIC GERANYLGERANYL DIPHOSPHATE SYNTHASE; PSY1, PHYTOENE SYNTHASE 1; PSY2, PHYTOENE SYNTHASE 2; PDS, PHYTOENE DESATURASE; ZDS, Ζ-CAROTENE DESATURASE; CRTISO, CAROTENOID LYCOPENE ISOMERASE; B-CYCLASE; LCY-E, CYC-B, LYCOPENE E-CYCLASE; CHROMOPLAST-SPECIFIC LCY-B, LYCOPENE CYCLASE; CRTR-B1, Β-RING HYDROXYLASE 1; CRTR-B2, Β-RING HYDROXYLASE 2; CYP97A, B-CAROTENE HYDROXYLASE P450; ZEP, ZEAXANTHIN EPOXIDASE; VDE, VIOLAXANTHIN DE-EPOXIDASE; NXS, NEOXANTHIN SYNTHASE; NCED, 9-CIS-EPOXYCAROTENOID DIOXYGENASE. TES: Tomato EST-SSR, TGS: Tomato Genome-SSR Map positions of the DNA markers used for genotyping are available at Kazusa DNA marker database (http://kazusa.or.jp/tomato/). Table S3. Genetic mapping of the pyp1-1 mutant. A; Homozygous genotype of Ailsa Craig. B, Homozygous genotype of Micro-Tom. H; Heterozygous genotype. S, Short arm. L; Large arm. cM, Centimorgan. M; Micro-Tom. A; Ailsa Craig. F1; F1 plants derived from a cross between Micro-Tom and Ailsa Craig. Number 1~24; F2 mapping populations between pyp1-1 and Ailsa Craig showing pale-yellow petals. DNA marker information including map position and primer information is available at the Kazusa DNA marker database (http://kazusa.or.jp/tomato/). 1 Refer to Shirasawa et al. (2010) in DNA Res. 17: 381-391. Table S4. Next-generation sequencing analysis and resulting coverage of the tomato genome. The number of reads produced by HiSeq 2000 and GA IIx are shown. Total reads represent total numbers of reads. Cleaned reads were produced using the SAMtools and used for mapping on tomato reference genome SL2.40. Mapped reads represent the number of reads mapped onto about 1.8 cM of tomato reference genome between SNP markers HMSNP2_551 and HMSNP8_1655, where the PYP1 locus was narrowed by map-based cloning (Chr1; position 78,889,698 80,651,803). Table S5. Identification of mutant-specific SNPs within the region demarcated by map-based cloning. Chr Position; physical position of mutation on the tomato reference genome (SL2.40). Reference (Heinz); Sequence of WT tomato Heinz genome (SL2.40). DP4; Four numbers correspond to numbered of forward ref alleles, reverse ref, forward non-ref, and reverse non-ref alleles, used in variant calling. Locus, mapped position for each pyp mutation. Mutation position; Exon indicates that homozygous SNPs found in each pyp1 mutant are located within the exon region, while intron indicates that the SNPs are located within the intron region. IGR, intergenic region. ITAG name, ITAG gene number defined in SL2.40. Effect of mutation; Silent indicates mutation causes synonymous mutation, while missense mutation causes non-synonymous mutation. Nonsense mutation introduces a stop codon. Table S6. Carotenoid contents of mature leaf and fruit in the WT and pyp1 mutants. Carotenoid contents in mature leaves and ripe red fruits were examined by HPLC analysis. The analysis was carried out in two distinct years (2010 and 2012).The mean values were calculated from at least five biological repetitions. Asterisks indicate significant difference compared to the WT (Student's t-test, P<0.05). DW; dry weight. Table S7. 500 MHz 1H-NMR data of tomato carotenoid esters in CDCl3. a d, Chemical Shift. b mult, Multiplicity (br, broad; d, doublet; dd, doublet of doublet; m, multiplet; s, singlet; t, triplet). c J, Coupling constant. Table S8. Scientific names and accession numbers of PYP homologues used for phylogenic analysis. Name of Figure S2; The name used for phylogenic tree analysis was listed here. Accession numbers were referred by NCBI website (http://blast.ncbi.nlm.nih.gov/Blast.cgi?CMD=Web&PAGE_TYPE=BlastHome), except for Moss, Lotus, Cucumber, Papaya, Green algae at Plant Genome Duplication Database (http://chibba.agtec.uga.edu/duplication/) and Melon from DDBJ database (http://www.ddbj.nig.ac.jp/).