Supporting information for online publication only Epioblasma
advertisement

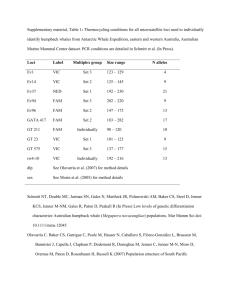
Supporting information for online publication only Epioblasma triquetra PCR conditions All 10 µl PCR reactions contained 1 µl (approximately 5 ng) DNA, 2l 5X PCR buffer containing 1.5 mM MgCl2 (Qiagen), an additional 0.35 mM MgCl2, 0.5 mM each dNTPs (BioShop), 0.23 mg/ml BSA (BioShop), 0.5 U Taq DNA polymerase (Qiagen) and the following concentrations of each forward (fluourescently labelled) and reverse primer: Multiplex 1: 0.2 µM LabC23 (AF512385); 0.25 µM Etr140 (DQ396406); 0.12 µM Ecap9 (AY650397); Multiplex 2: 0.25 µM Ecap5 (AY650393); 0.15 µM LabD213 (AF512398); Multiplex 3: 0.1 µM LabC2 (AF512384); 0.3 µM Etr90 (DQ396403); 0.35 µM LabD206 (AF512395); Multiplex 4: 0.15 µM Etr114 (DQ396404); 0.25 µM Etr124 (DQ396405); 0.1 µM LabC24 (AF512386); 0.45 µM Etr145 (DQ396407); 0.1 µM Ecap6 (AY650394); Multiplex 5: 0.35 µM each of Etr187 (DQ396408) and LabD111 (AF512395). Total reaction volumens were made to 10 l with PCRgrade ddH20 (BioShop). Amplification consisted of 2 min at 94oC followed by 40 cycles of 94oC (40 s), an annealing temperature of 58oC (40 s), 72oC (60 s), with a final extension at 72oC (5 min). Due to inconsistent amplification success loci Ecap9, LabC2, LabC24, and Etr187 were excluded from all analyses. Quadrula quadrula PCR conditions PCR reactions for locus QfA103 contained 1 µl (approximately 5 ng) DNA, 2l 5X PCR buffer containing 1.5 mM MgCl2 (Qiagen), an additional 0.5 mM MgCl2, 0.2 mM each dNTPs (BioShop), 0.2 mg/ml BSA (BioShop), 0.5 U Taq DNA polymerase (Qiagen), 0.1 µM forward primer with an M13 tail (TGT AA ACG ACG GCC AGT) at the 5’ end, 0.4 µM reverse primer, and 0.4 µM fluorescently labelled M13 probe (Schuelke 2000) in a total volume of 10 µl. PCR reactions for the remaining loci were the same except they contained 0.4 µM each of a fluorescently labeled forward primer and 0.4 µM reverse primer in a total volume of 10 µl. Amplification consisted of 10 min at 94oC followed by 40 cycles of 94oC (45 s), 60 s at the optimal annealing temperature, 72oC (60 s), with a final extension at 72oC (5 min). Annealing temperatures (Genbank accession numbers) were as follows for each locus: 48oC for QfA103 (FJ785629); 51oC for QfD102 (FJ85635); 55oC for QfA112 (FJ85630) and QfA130 (FJ785631); and 59oC for QfC114 (FJ85634) and QfC4 (FJ85632). Lampsilis fasciola PCR conditions All 10 µl PCR reactions contained 1 µl (approximately 5 ng) DNA, 2l 5X PCR buffer containing 1.5 mM MgCl2 (Qiagen), an additional 0.35 mM MgCl2, 0.5 mM each dNTPs (BioShop), 0.23 mg/ml BSA (BioShop), 0.5 U Taq DNA polymerase (Qiagen) and the following concentrations of each forward (fluourescently labelled) and reverse primer (Genbank accession number): Multiplex 1: 0.1 µM Ecap1 (AY650389); 0.15 µM Ecap5 (AY650393); Multiplex 2: 0.1 µM each of Etr114 (DQ396404), LabC23 (AF512385), and LabC24 (AF512386); Multiplex 3: 0.1 µM LabD206 (AF512395); 0.075 µM LabD213 (AF512398); Multiplex 4: 0.1 µM LabC2 (AF512384); 0.2 µM LabD111 (AF512395). Total reaction volumens were made to 10 l with PCR-grade ddH20 (BioShop). Amplification consisted of 2 min at 94oC followed by 40 cycles of 94oC (40 s), an annealing temperature of 58oC (40 s), 72oC (60 s), with a final extension at 72oC (5 min). Due to inconsistent amplification success locus LabC2 was excluded from all analyses. Amblema plicata PCR conditions All 10 µl PCR reactions contained 1 µl (approximately 5 ng) DNA, 2l 5X PCR buffer containing 1.5 mM MgCl2 (Qiagen), an additional 0.5 mM MgCl2, 0.5 mM each dNTPs (BioShop), 0.2 mg/ml BSA (BioShop), 0.25 U Taq DNA polymerase (Qiagen), 0.1 µM of each forward primer with an M13 tail (TGT AA ACG ACG GCC AGT) at the 5’ end, 0.3 µM of each reverse primer and 0.4 µM fluroescently labelled M13 probe (Schuelke 2000). Total reaction volumens were made to 10 l with PCR-grade ddH20 (BioShop). Amplification consisted of 10 min at 94oC followed by 35 cycles of 94oC (45 s), 60 s at the optimal annealing temperature, 60 s at 72oC, with a final extension at 72oC (5 min). Annealing temperatures (Genbank accession numbers) were as follows for each locus: 58oC for Anec103 (JF719044); 51oC for Anec114 (JF719045); 48oC for Anec126 (JF19049); 60oC for Aned103 (JF719052); 56oC for Aned104 (JF19053); 54oC for Aned108 (JF19055) and Aned140 (JF19059); and 50oC for Aned126 (JF19056). Thames Grand Ausable Sydenham Supplemental Table 1: Summary of samples for the six mussel species, showing sampled rivers, sites, and number of genotyped loci, and local sample sizes (numbers of sampled individuals) for each species and location. Sites with n<10 were removed from all population-based analyses to avoid bias, but were included for individual-based assignments. Site locations are shown in Figure 1. E. triquetra P. fasciolaris Q. quadrula L. fasciola L. costata A. plicata River Site (11 loci) (9 loci) (6 loci) (8 loci) (11 loci) (8 loci) 1 2 3 4 5 Total 6 7 8 Total 9 10 11 12 13 14 Total 15 16 17 18 19 20 Total Grand Total 23 4 7 3 27 64 30 0 0 30 0 0 0 0 0 0 0 0 0 0 0 0 0 0 17 14 16 9 49 105 0 25 25 50 0 0 0 0 0 0 0 0 0 0 0 0 0 0 68 38 54 23 47 230 0 0 0 0 20 19 2 21 0 0 62 21 25 67 0 0 0 113 0 0 0 0 0 0 0 0 0 0 0 0 0 0 18 38 56 0 0 0 32 50 37 119 19 4 9 10 23 65 24 9 25 58 0 0 0 0 20 19 39 4 2 0 18 28 27 79 11 9 10 7 0 37 25 10 20 55 0 0 0 0 0 0 0 0 0 0 0 0 0 0 94 155 405 175 241 92