tpj12940-sup-0007-Legends
advertisement

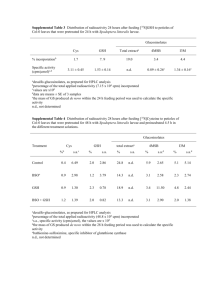
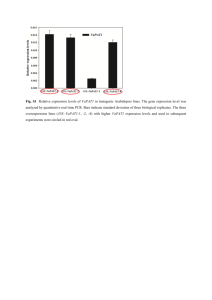
Supporting Information Legends Figure S1. Effects of exogenous GSH on drought and salt tolerance. For drought and salt stress treatment, 10-d-old wild-type plants were incubated in water containing GSH at 22°C under 16h-light/8-h-dark conditions for 2 weeks. Newly prepared solution was supplied every other day. Mock indicates plants growing in water solution only. Each treatment consisted of three pots with 20 plants in each pot. Five independent experiments were performed with similar results. Figure S2. Expression levels of GSH1 in transgenic plants. (a) GSH1 expression in 4-week-old RD29A:GSH1 (line 7, 12, and 15) transgenic plants under normal condition (Control) or withholding water for 5 d (Drought). (b) GSH1 expression in 35S:GSH1 transgenic plants in normal condition. For both experiments, total RNA was extracted from whole plants and 1μg of total RNA from each sample was used for reverse transcription. Figure S3. Drought tolerance of pad2-1 mutants. Six plants for each line were planted on each pot for drought stress test. Drought tolerance of wild-type (WT) and pad2-1 mutants after withholding water for 12~16 d and rehydration for 4 d. Figure S4. GSH and GSSG contents in WT plants treated with 50 μM BSO. Ten-d-old plants grown in soil under long-day (LD) condition were watered with or without 50 μM BSO every other day for 14 d. Biological triplicates were averaged. Bars indicate SE (*P < 0.05, Student’s t-test was performed of the values of GSH-treated plants against control plants.). Figure S5. Determination of GSH (a) and GSSG (b) levels in shoot and root under 8 h and 24 h of 100 μM GSH treatment. Values are presented as means ± SD from five samples for each time point. Two independent experiments were performed with similar results. *P<0.1, Student’s ttest was performed of the values of GSH-treated plants against control plants. (c) Cellular glutathione redox in the roots and shoots of Col-0 expressing GRX1-roGFP2 treated with or without GSH. Figure S6. GSH1 expression (a) and total GSH levels (b) in Col-0 and RD29A:GSH1 transgenic seedlings after salt stress treatment. Values are presented as means ± SD from five samples for each time point. Two independent experiments were performed with similar results. *P<0.1, Student’s t-test was performed of the values of salt stress-treated plants against control plants. Figure S7. Salt stress-induced GSH1 enhance translation activity. (a) Polysome profiles after sucrose gradient centrifugation of 10-d-old RD29A:GSH1 and Col-0 seedlings treated with control medium or 150 mM NaCl-containing medium for 0, 3, and 6 h. (b) Bar graph showing the ribosome loading efficiency in 10-d-old RD29A:GSH1 and Col-0 seedlings treated with control medium or 150 mM NaCl-containing medium for 0, 3, and 6 h. Values are mean percentages ± SE from three biological replicates. *P < 0.05, Student’s t-test. Figure S8. Confirmation of increased ribosome occupancy by qRT–PCR. Changes in polysomebound mRNA (a) and steady-state (b) abundance between GSH-treated and control samples (open square) with standard deviations were calculated from three technical repeats of one representative biological repeat. Transcriptome data obtained from Microarray hybridization were also plotted (filled square). Figure S9. Experimental validation of GSH-responsive genes regulated at the translational level. (a) Immunoblotting was used to detect indicated proteins in different tissues at different times. Endogenous α-tubulin (a-tubulin) was used as a loading control. The number below each blot represents the relative level of the indicated protein. The level in samples treated without GSH was arbitrarily set to 1. Samples of 24 h-root and 24 h-shoot were loaded on separate gels. (b) A table showing the fold change values of validated genes at PL level obtained from microarray analysis. Figure S10. Changes in polysome-bound mRNA and steady-state abundance between Col-0 and RD29A:GSH1 seedlings treated with salt stress for 3 h and 6 h by qRT-PCR. Standard deviations were calculated from three technical repeats of one representative biological repeat. Figure S11. Categorization of genes regulated by GSH treatment at the mRNA or protein levels. (a) and (c), The upregulated genes at 8R (a) and 24S (c) (with ≧ two-fold changes at mRNASS or mRNAPL levels) were categorized into three groups, preferentially regulated at the steady-state mRNA level (RNA), the polysome-bound mRNA level (Protein) or both (RNA+Protein). Extreme red and green colors indicate six-fold upregulation and 0.4-fold downregulation, respectively. (b) and (d), DAVID functional clustering enrichment score (ES) of the highly enriched GO terms in respective category of 8R- (b) and 24S- (d) regulated genes. Figure S12. Cluster analysis of the differentially expressed genes at the steady state level after 8 h and 24 h of GSH treatment. R, root; S, shoot. Figure S13. Pro content in GSH-treated plants (a) and 35S:GSH1 (OE7-5 and OE13-6) transgenic plants (b). Data are presented as means and SE of three replications. *P<0.1, Student’s t-test was performed of the values of GSH-treated plants against control plants (a) and mutant plants against WT plants (b). Values are presented as micromole per gram of dry weight for each sample. Total Pro was prepared from 10-d-old Arabidopsis grown on MS agar plates. Pro contents were also measured after transferring 10-d-old seedlings onto medium containing 100 μM GSH for 24 h. WT, wild type. Figure S14. Heat map showing the differential expression of JA-responsive genes translationally regulated after 8 h and 24 h of GSH treatment. Extreme red and green colors indicate two-fold upregulation and downregulation, respectively. Figure S15. Heat map showing the differential expression of auxin-responsive genes regulated translationally after 8 h and 24 h of GSH treatment. Extreme red and green colors indicate twofold upregulation and downregulation, respectively. Figure S16. Heat map showing the differential expression of ABA-responsive genes regulated translationally after 8 h and 24 h of GSH treatment. Extreme red and green colors indicate twofold upregulation and downregulation, respectively. Figure S17. Effect of exogenous GSH on ABA accumulation in root and shoot tissues. Ten-d-old wild-type plants were transferred to half-strength MS medium containing GSH (0 and 100 μM) 8 and 24 h. Values represent means ± SD from three samples. Three independent experiments were performed with similar results. *P<0.1, Student’s t-test was performed of the values of GSHtreated plants against control plants. Supplementary experimental procedures. Data Set 1. Genes with ≥ 2-fold changes at the mRNASS level. Data Set 2. Genes with ≥ 2-fold changes at the mRNAPL level. Data Set 3. Expression ratios of genes regulated at 8R and 24S organized by k-means clustering and GO analysis results. Data Set 4. Gene lists and GO analysis results of genes regulated in 8R and 24S at the mRNAPL level.