Part 1: Type 2 Diabetes Mellitus Phenotype Algorithm
advertisement

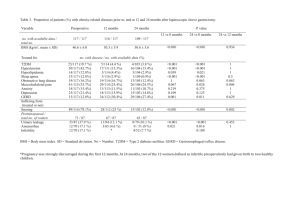
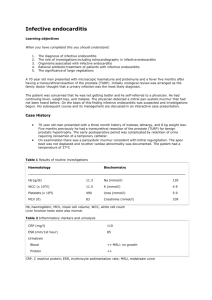
Desiderata for Computable Representations of Electronic Health Records-Driven Phenotype Algorithms Supplemental Document (Appendix) Part 1: Type 2 Diabetes Mellitus Phenotype Algorithm This section provides a high-level overview of the eMERGE Type 2 diabetes mellitus (T2DM) algorithm [1] for extracting T2DM cases from the EHR (please see https://phekb.org/phenotype/type-2-diabetes-mellitus for further details). The case selection algorithm requires specific patient-level data elements to be extracted from the EHR, including diagnoses, lab results, medication orders, and physician encounter dates. In particular, the following data elements (defined with standardized concept lists, as Desideratum 7) are required: Counts of type 1 diabetes mellitus (T1DM) ICD-9 code assignment dates by diagnostic source Counts of T2DM ICD-9 code assignment dates by diagnostic source T1DM medications (i.e., insulin & Symlin) order or prescription dates – at least the earliest date of Rx T2DM medications (metformin & other sulfonylureas, biguanides, etc.) order or prescription dates – at least the earliest date of Rx Fasting blood glucose lab values – at least the maximum value Random blood glucose lab values – at least the maximum value HBA1c lab values – at least the maximum value For this algorithm, the following definitions and abbreviations apply: Abnormal lab – An abnormal lab value is defined as one of the following: – Random glucose > 200 mg/dl – Fasting glucose ≥ 125 mg/dl – Hemoglobin A1c ≥ 6.5% Physician entered diagnosis – A physician entered diagnosis code is one that is derived from encounter or problem list sources only (excludes diagnoses entered into billing systems). A flowchart expressing the logic of the T2DM case selection algorithm is shown in Figure 1. Figure 1 (same as main text): Phenotype algorithm for identifying type 2 diabetes mellitus (T2DM) from electronic medical records (EMR, or EHR). T1DM: type 1 diabetes mellitus; Dx: diagnoses, defined as recorded using International Classification of Diseases, 9th Revision (ICD-9) codes; med: medication; physcn: physicians; Rx: prescriptions. (From PheKB.org) The T2DM algorithm has been implemented as a workflow inside of the Konstanz Information Miner (KNIME) data analytics platform. The workflow takes as input a comma-separated value (CSV) file, with each row corresponding to a set of patient-level variables (as Desideratum 2). Set operations (as Desideratum 4) are mostly performed at the relational level in generating patient-level values, some of which are counts and dates of first appearance (as aggregative operations in Desideratum 5). Figure S2 shows the top-level workflow that implements the algorithm. Figure S3 show the input data in tabular format – each row contains a set of data for one patient, each column is a data item with a specific data type. Figure S4 shows the configuration window for the final Rule Engine node in the workflow. This node implements the logic depicted by the flowchart diagram in Figure 1 (as Desideratum 5). One of the rule paths for cases requires that the (first) oral hypoglycemic medications (as preferable T2DM medications) should start before start of the (first) insulin treatments (as preferable T1DM medications), as Desiderata 6. Figure S2: T2DM implementation as a KNIME workflow. Figure S3: Table of deidentified* input data for KNIME workflow. Each row contains a set of patient-level variables (*patient IDs are removed and the dates are randomly shifted by a different random number for each patient for protection of patient privacy). Figure S4: Configuration window for the final Rule Engine node in Figure S2. This node encodes test values of variables and returns either “case” or “unknown” for each patient. This logic implements the paths through the flowchart in Figure 1. Part 2: Examples of complex counting rules and scoring rules (for Desideratum 5, aggregative operations. Modified Duke Criteria (a counting rule) [2]: Table A: Definitions of criteria Major criteria 1. Positive blood cultures for IE Typical microorganism for infective endocarditis from two separate blood cultures: Viridans streptococci Streptococcus gallolyticus (formerly S. bovis), including nutritional variant strains (Granulicatella spp and Abiotrophia defectiva) HACEK group: Haemophilus spp, Aggregatibacter (formerly Actinobacillus actinomycete comitants), Cardiobacterium hominis, Eikenella spp, and Kingella kingae Staphylococcus aureus Community-acquired enterococci, in the absence of a primary focus; OR Persistently positive blood culture, defined as recovery of a microorganism consistent with IE from: Blood cultures drawn more than 12 hours apart OR All of three or a majority of four or more separate blood cultures, with first and last drawn at least one hour apart Single positive blood culture for Coxiella burnetii or antiphase I IgG antibody titer >1:800* 2. Evidence of endocardial involvement Positive echocardiogram for IE TEE recommended in patients with prosthetic valves, rated at least "possible IE" by clinical criteria, or complicated IE [paravalvular abscess]; TTE as first test in other patients Definition of positive echocardiogram Oscillating intracardiac mass, on valve or supporting structures, or in the path of regurgitant jets, or on implanted material, in the absence of an alternative anatomic explanation OR Abscess OR New partial dehiscence of prosthetic valve New valvular regurgitation Increase in or change in preexisting murmur not sufficient Minor criteria 1. Predisposition: predisposing heart condition or intravenous drug use 2. Fever: 38.0°C (100.4°F) 3. Vascular phenomena: major arterial emboli, septic pulmonary infarcts, mycotic aneurysm, intracranial hemorrhage, conjunctival hemorrhages, Janeway lesions 4. Immunologic phenomena: glomerulonephritis, Osler's nodes, Roth spots, rheumatoid factor 5. Microbiologic evidence: positive blood culture but not meeting major criterion as noted previously (excluding single positive cultures for coagulase-negative straphylococci and organisms that do not cause endocarditis) OR serologic evidence of active infection with organism consistent with IE 6. Echocardiographic minor criteria eliminated IE: infective endocarditis; TEE: transesophageal echocardiography; TTE: transthoracic echocardiography. Table B: Definitions of diagnosis Definite IE Pathologic criteria Microorganism: demonstrated by culture or histology in a vegetation, or in a vegetation that has embolized, or in an intracardiac abscess OR Pathologic lesions: vegetation or intracardiac abscess, confirmed by histology showing active endocarditis Clinical criteria Using specific definitions listed in Table B: 2 major criteria OR 1 major and 3 minor criteria OR 5 minor criteria Possible IE 1 major criterion and 1 minor criterion OR 3 minor criteria Rejected IE Firm alternate diagnosis for manifestations of endocarditis OR Resolution of manifestations of endocarditis, with antibiotic therapy for four days or less OR No pathologic evidence of infective endocarditis at surgery or autopsy after antibiotic therapy for four days or less Does not meet criteria for possible infective endocarditis, as above IE: Infective Endocarditis. CHA2DS2_VASc Scores for anti-coagulation therapy in atrial fibrillation (a scoring rule) [3]: CHA2DS2-VASc definition Score Congestive heart failure 1 Hypertension 1 Age ≥ 75 years 2 Diabetes mellitus 1 Stroke/transient ischemic attack/thromboembolism 2 Vascular disease (prior myocardial infarction, peripheral artery disease, aortic plaque) 1 Age 65 to 74 years 1 Sex category (ie, female sex) 1 Maximum score CHA2DS2-VASc score Unadjusted ischemic stroke rate (% per year) 0 0.2% 1 0.6% 2 2.2% 3 3.2% 4 4.8% 5 7.2% 6 9.7% 7 11.2% 8 10.8% 9 12.2% 9 Part 3: Implementation of PheRM desiderata in PhEMA The Phenotype Execution Modeling Architecture (PhEMA) project is a multiinstitutional collaboration in developing a common PheRM (Phenotype Representation Model (PheRM) and its infrastructure to apply PheRM to multi-center EHR-based biomedical research (e.g., eMERGE network). During the ongoing implementations in PhEMA, these desiderata (D1 ~ D10) have been and are being constantly tested for practicality. The first PhEMA application being developed is an authoring tool, and it delivers two services (as D3): a graphical interface for developing, storing, and exporting humanunderstandable representations, and a validation service to ensure that authored algorithms are executable by translating them into QDM, KNIME workflows, SQL, and Fast Healthcare Interoperability Resources (FHIR) queries. The second PhEMA application being developed is a translator, which converts QDM (a mostly rule-based representation, as D5, and rich in temporal comparisons, as D6) into KNIME workflows (an executional environment based on relational tables, as proposed in D4). Both of these efforts are using VSAC services and standard HL7 terminologies (as D7). As a further step, the QDM to KNIME translator is able to construct queries against a local IDR by either directly matching concept identifiers or converting a concept list into regular expressions for searching. Both PhEMA and eMERGE are actively adopting a KNIME solution to dispatch phenotype algorithms for real-world research projects. As previously described, KNIME workflows allow bridging to external packages (e.g., Java, Python, R, Perl) to perform statistical classifications and project specific NLP (as D9). KNIME workflows are also amenable to adapt to local IDR with last-step simple queries (as D2). As a significant executable solution, KNIME provides the breadth of a full collection of generic computational tools to bridge the limitations of state-of-the-art PheRM implementations, ensuring that phenotyping projects will not be hindered from using our PheRM due to lack of features that could not be foreseen as being necessary or are not yet available. References for the Appendix: 1 Kho AN, Hayes MG, Rasmussen-Torvik L, et al. Use of diverse electronic medical record systems to identify genetic risk for type 2 diabetes within a genome-wide association study. J Am Med Inform Assoc JAMIA 2012;19:212–8. doi:10.1136/amiajnl-2011-000439 2 Li JS, Sexton DJ, Mick N, et al. Proposed modifications to the Duke criteria for the diagnosis of infective endocarditis. Clin Infect Dis Off Publ Infect Dis Soc Am 2000;30:633–8. doi:10.1086/313753 3 Friberg L, Rosenqvist M, Lip GYH. Evaluation of risk stratification schemes for ischaemic stroke and bleeding in 182 678 patients with atrial fibrillation: the Swedish Atrial Fibrillation cohort study. Eur Heart J 2012;33:1500–10. doi:10.1093/eurheartj/ehr488