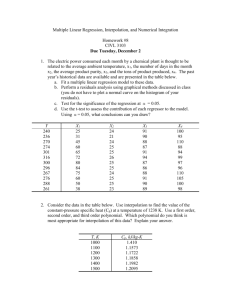
Matrix Algebra Rows x Columns Scalar matrix is a diagonal matrix
advertisement

Matrix Algebra Rows x Columns Scalar matrix is a diagonal matrix (all zeroes except for diagonal) with all the same value. identity has ones on diagonal. Transpose mirrors the matrix, then turns it 90 degrees to the right. #rows become # columns. Multiplication: The interior dimensions must be the same. (3 x2 and 2 x 3 share inner dimensions (2)). The resulting matrix will be the outer dimensions (3 x 3). Row dot multiplied by column. Fo through all columns. This makes up the first row in the resulting matrix. Division: Multiply the reciprocal (the inverse). Division is equivalent to premultiplication by the inverse of a matrix. B/A = A-1B Matrix Inverse Steps 𝐴−1 = 1 𝑎𝑑𝑗𝐴 |𝐴| 1) calculate determinant = |A| a. the product of main diagonal elements minus the product of minor diagonal elements. i. think about a covariance matrix. If shared variance (covariance) is larger, then the determinant will be small. ii. the higher the related predictors, the lower the determinant. 1. perfectly collinear will give a determinant of 0. iii. So, we want to deal with X’X to get a square matrix, first. b. is a scalar. 2) find the adjugate a. mirror the diagonal b. switch the signs of the off-diagonals 3) calculate inverse If you are distributing a transpose or an inverse, then be sure to mirror the order! If you want a mean, then you can multiply a row vector by a column vector of same length made up of ones and the multiply by scalar (1/n). To get sum of squares, just do X’X. This multiplies each element by itself and sums up. To get the sum of a variable, square… do X’ multiplied by a unit vector You can get sums by just multiplying by a unit vector. SSCP for an entire matrix is X’X-1/nX’EX We can now use this info in order to bring our regression equation into matrix form. We need to go through the same logic we used for deriving our OLS equation. Particularly that we want to minimize our residuals. First thing we need to do is include the intercept in our X matrix by adding a column of 1s to the front. Everything else makes sense as the stored form of our variables before. If our predicted y is equal to Xb, then it follows that residuals are y-XB, so that (squared) is what we want to minimize. We format this as e’e. We know that: e=y-XB, so e’ will be (y-Xb)’ but we need to distribute the transpose and switch order of multiplied variables: e’=(y’-b’X’) & e=(y-Xb) So distributing, to get e’e would get: y’y-y’Xb-b’X’y+b’X’Xb Our middle terms(y’Xb-b’X’y) are scalars and transposes of eachother. y’ will be 1 x n. b will also have 1 row. So, the end result will be 1 number. Additionally, they are transposes of eachother. Scalars that are transposes of eachother are equal to eachother, so we can simplogy by duplicating the second term and dropping the 3rd. This gives us: y’y-2y’Xb+b’X’Xb This is what we now want to differentiate in order to minimize in respect to each of the variables of interest (mainly b) and set that to 0. The end of that process is: b=(X’X)-1X’y’ This is the matrix expression for producing estimates of the regression coefficients. Onve we have the bs we can do Xb to get our predicted y. Which, if we think about it is equivalent of timing X by our matrix regression form: Xb = X(X’X)-1X’y This is the form of Hy (the hat matrix) which is basically the weighted Xs that make up b. It will always be of form nxn. Predicted values, thus, are a linear combination of observed values. We can also use matrix math to get key regression data points, like SSy: 1 𝑆𝑆𝑦 = 𝑦 ′ 𝑦 − 𝑦′𝐸𝑦 𝑛 SSres is the same as e’e SSreg is just SSy-SSres which is the same as: 1 𝑆𝑆𝑟𝑒𝑔 = 𝑏 ′ 𝑋 ′ 𝑦 − 𝑦′𝐸𝑦 𝑛 Remember that R2 is always the ratio of SSreg over SSy Remember the formula for F: 𝑅2 𝑘 𝐹= (1 − 𝑅 2 ) (𝑛 − 𝑘 − 1) If we want to know about the standard error (and subsequent significance) of our b values then we need to get a variance/covariance matrix: 2 𝑠(𝑏) = 𝑀𝑆𝑟𝑒𝑠 (𝑋 ′ 𝑋)−1 This will yield a [k+1] x [k+1] matrix where the diagonal elements are the variance of b’s. So, square root of them are the standard errors. The off-diagonals are the sampling covariances. Then you do a t-test of b value over its standard error. We can also use constrast codes in order to compare the linear combinations of bs. First, as usual..we need to combine our errors. 2 𝑠𝑏1−𝑏2 = 𝑣𝑎𝑟(𝑏1) + 𝑣𝑎𝑟(𝑏2) − 2𝐶𝑜𝑣(𝑏1, 𝑏2) OR (where c is a vector of contrast codes) 2 2 𝑠𝑏1−𝑏2 = 𝑐 ′ 𝑠(𝑏) Technically, we don’t need raw data to run a regression. We just need the correlations, the means, the variances, and the SDs (and n). The matrix expression is technically just computed regression coefficients from correlations. Technically: 𝛽 = 𝑅𝑋𝑋 −1 𝑟𝑋𝑌 Then we can just find our bs by the typical transformation: 𝑠 𝑏 = 𝛽 𝑠𝑌 and 𝑏𝑜 = 𝑌̅ − ∑ 𝑏𝑋̅ 𝑋 If we have multicolinearity, we can’t invert the data matrix. We can detect multicolinearity through: Tolerenace(which we want to be below .16 2 𝑇𝑜𝑙𝑒𝑟𝑎𝑛𝑐𝑒 = 1 − 𝑅1.2…𝑘 Variance inflation factor(which we want to be above 6) 𝑉𝐼𝐹 = 1 2 1 − 𝑅1.2…𝑘 Basically, there terms speak to how much variance remains in a variable after partialling out all the other variables. If we have multicolinearity, then, by nature of the equation for a standard error: 𝑠 𝑀𝑆𝑟𝑒𝑠 𝑏𝑦1.2 =√ 2 ) 𝑆𝑆𝑋1 (1−𝑟12 So, since the denominator is 1-correlation, then a higher correlation, will make a lower denominator and, in turn, a larger standard error. This large standard error will make it hard to find significance for individual predictors and could give us “bouncing betas” where variations in our predictor set may cause large changes in the magnitude of the coefficients. Predictor subsets (cross-validation) could really mess this up. We can begin to drop predictors that have the least tolerance (their shared variance will go to the remaining predictors. We can also combine predictors into composites. We can also center (to reduce non-essential colinearity with powered / interaction vectors). We can also add cases that break the pattern of multicolinearity. We only think of correlations as high because they are close to 1. If we artificially increase our correlation matrix to where the diagnonals are not 1, but increased by a constant, then the correlations are no longer relatively high. This is the basis of ridge regression: A small degree of bias in coefficients is traded for much smaller variances(standard errors). Diagnostics We can determine outliers with our hat matrix. The diagonal values will tell us the leverage that each of our X variables has. We has previously defined leverage as: 1 (𝑋 − 𝑋̅)2 𝑙𝑒𝑣𝑒𝑟𝑎𝑔𝑒 = ℎ𝑖𝑖 = [ + ] 𝑛 𝑆𝑆𝑋 The average leverage value is k/n, so its problematic is our leverage is >2*k/n We can also get Mahalanobis Distance which is (n-1)*leverage An outlier is a point that is discrepant in terms of its Y values, given its X values. So we could see this based on the residuals for that particular Y point. Particular if we standardize the residuals. We can’t just divide each residual by square root of MSerror. At the population level we need to assume that epsiolons are uncorrelated and have constant variance for all values of X. However, the residuals are correlated and their variance changes depending on X: variance of residual is large close to the mean and small far from it. We can get an internally studentized residual that takes this into account by using our leverage value when all the data points are in the model: 𝑠𝑒𝑒𝑖 = √𝑀𝑆𝑟𝑒𝑠 (1 − ℎ𝑖𝑖 ) However, if a point is an extreme outlier, then it might be pulling the regression line towards it and thus decreasing the residual for that point. We could instead just delete that point, run the regression (on n-1 cases) and evaluate the deleted point relative to the new line. This would be our externally studentized residual(Y-Yhat) where Yhat is the predicted value of Y when Y’s Xs were not in the model. Remember that not anomalous points are outliers. Only large leverage and large residuals mean that the point is influential on data. Thus, 𝑖𝑛𝑓𝑙𝑢𝑒𝑛𝑐𝑒 = 𝑑𝑖𝑠𝑡𝑎𝑛𝑐𝑒 ∗ 𝑙𝑒𝑣𝑒𝑟𝑎𝑔𝑒 We can measure this influence directly with Cook’s D. Basically, it is the sum of squared changes in the regression coefficient that would occur if that individual were removed from the data and the analysis stream. A value over 1 is considered unusual. We can see it in statistical format as follows: 𝐶𝐷𝑖 = 𝑒2 𝑙𝑒𝑣𝑒𝑟𝑎𝑔𝑒 ∗ 𝑘𝑀𝑆𝑟𝑒𝑠 (1 − 𝑙𝑒𝑣𝑒𝑟𝑎𝑔𝑒)2 If this CD value is greater than 4/(n-k-1), then that point is highly influential. We can also look directly at the influence of a point on the fitted values (showing how much one point changes the fitness of a line). 𝐷𝐹𝐹𝐼𝑇𝑆𝑖 = 𝑌̂𝑖 − 𝑌̂−𝑖 √𝑀𝑆𝑟𝑒𝑠(−𝑖) ℎ𝑖𝑖 We want: 𝐷𝐹𝐹𝐼𝑇𝑆 < 2√ 𝑘+1 𝑛−𝑘−1 We can also measure how much our betas are getting affected: 𝐷𝐹𝐵𝐸𝑇𝐴𝑏1 = 𝑑𝑖𝑓𝑓𝑒𝑟𝑒𝑛𝑐𝑒 𝑖𝑛 𝑏𝑒𝑡𝑎𝑠 𝑤𝑖𝑡ℎ 𝑎𝑛𝑑 𝑤𝑖𝑡ℎ𝑜𝑢𝑡 𝑎 𝑐𝑎𝑠𝑒 √𝑀𝑆𝑟𝑒𝑠(𝑤𝑖𝑡ℎ𝑜𝑢𝑡𝑐𝑎𝑠𝑒) 𝑐 c would be the diagonal element of (X’X)-1 which would basically be b1 and X1, thus making the denominator the standard error of the betas. We want DFBETA to be greater than 1. The cases might also have an influence on the standard errors of our predictors. To assess this, we can look at COVRATION which is a sampling variance for all the coefficients (ratio of volumes of deleted and full data confidence regions). An observation that increases precision produces a large COVRATIO (which will >1 since the null is 1 – no change). If there are several similar outliers, they may be a subpopulation. Don’t forget about winsorizing (taking the anomalous value and making it the same value as the second highest value). We can get a measure called reliability by projecting a true score theory onto the population: True score would be X = T+E where X is observed, T is actual, and E is a random error. Then, we can look at the sigma of that true score and compare it to the actual sigma: 𝑟𝑒𝑙𝑖𝑎𝑏𝑖𝑙𝑖𝑡𝑦 = 𝜎𝑇2 𝜎𝑋2 This speaks to measurement error. If there is high measurement error, then our criterion(Y) is messing up our standard errors and thus attenuate the estimate of the b coefficient. To address this, we can make a correlation matrix and then adjust it (attenuate it): ∗ 𝑟12 = 𝑟12 √𝑟11 𝑟22 How can we really detect violations, though? we can plot and observe heteroscedasticity (where ranges of our data differ from other ranges in terms of their residuals). A plot that has residuals that “megaphones” (opening wider) to the right means that the standard errors are downwardly biased. We would transform Y down the ladder of reexpression in this case. Vice Versa. We can weight our betas by the sigma at each value of X (assuming our X can only take on a limited range of values and there are multiple Xs at each range). We need to make a weight matrix W, where 1 𝜎2 This will give small weights to those with larger error variances and vice versa. 𝑊= Thus, now b will be equal to: 𝑏(𝑤𝑙𝑠) = (𝑋 ′ 𝑊𝑋)−1 𝑋′𝑊𝑦 If our data is across time, then we can plot across time and look for weird bumps. If its too smooth it might be a positive autocorrelation. Too jagged, probably negative autocorrelation. For clustered data, you’ll want to calculate the intraclass correlation coefficient (ICC). We can plot our ordered residuals against expected magnitudes of residuals from a true normal distribution in a Q-Q plot. If its paraboling up, then we have a positive skew. Negative if parabola is opening down. If positive then negative then positive curvilinear then the tails are heavy. If negative then postivie then engative, then the tails are light but the peakedness is high (high kurtosis). If these errors are not normal, then OLS is still unbiased, but the t-tests are invalid. We can use Box-Cox to find the right transformation of Y that will minimize SSres. While we cant test for it, violate that the expected error term is 0 overestimates our weights. The intercept will be biased. Also, if the relationship between our X and our residuals is not zeros, then our bs will be biased. This is usually seen with model misspecification. Because if we have 3 variables, but 4 and 5 are left out, then their contribution to the potential prediction is all in error and then that error would not necessarily be even across all values of each of the Xs. We would especially see this if the omitted predictor is correlated with an included one, because then the error term would be correlated with the predictors. This can extend to whether or not to include power transforms. non-monotonic relationship…add powered vector terms monotoninc…add powered vector terms or transform X or Y via bulging rule and ladder of reexpression. We can make partial regression plots by plotting residuals from the first equation against the part of X2 that is unrelated to X1 (i.e residuals from predicting X2 from X1). Regression residuals are uncorrelated with predictors (or supposed to be). Logistic Regression If our dependent variable is 1 or 0, we can use predictors to determine the probability of Y being 1 or being 0. Part of this set up gives us non-normal errors (since errors can only be within 1 of eachother). Furthermore, error variance is non-constant since a dichotomous Y will involve X thus making variances different for different X values. Logistic regression is just an extension of odds which are just ratios (not proportions). You would say that odds that numerator thinks DV. Odds are asymmetric about 1: the distance between 0 and 1 and the distance between 1 and positive infinity express the same relation.So, we can correct for this asymmetry by taking the natural log. If we do this, the odds ratios within a category will sum to 0 (negatives of eachother). With natural logs, we are asking…to what power must we raise e in order to get X? The natural log of the odds is called the logit. If we have multiple conditions where we are calculating odds, we can get an odds ratio. The odds ratio for the variable of interest would be the categories odd ratio over the other categories odd ratio. This measures the association between binary X and Y variables. The null value would be 1. The multiplicative interpretation is that by change the category (or source), we can change the original ratio. We can say things like…by being in category A, someone is Odds Ratio times more likely to think DV than if they were in category B. We can also say that the odds of the DV increase by OddsRatio*100 – 100 %. Once again we can address asymmetry by taking the natural log. We can also deal with proportions, which has to do with the ratio of one variable with the sum of multiple. if there are 10 people that think something is biased and 10 that think its unbiased, then the porbabliity of someone thinking it is biased is 50%, since there are 20 total people. We can get odds by: 𝑂𝑑𝑑𝑠 = 𝑃 1−𝑃 We want a curve that can fit the distribution of odds (something that asymtotes at 0 and 1)…sigmoid. 𝑒 𝑏0+𝑏1𝑋 𝑃= 1 + 𝑒 𝑏0+𝑏1𝑋 This is the formula for a logistic curve. Technically, we can reduce this to be: ln ( 𝑝 ) = 𝑏0 + 𝑏1𝑋 1−𝑝 Maximum likelihood will help us understand which estimate of parameter values will give us back the data we already have. We want to maximize the likelihood function. We do this with a guided trial-and-error process to continually update our paramaters (gradient descent?). We can get an estimate of maximum likelihood known as deviance: Deviance = -2 * log likelihood This will be a positive number that measures our lack of fit and is useful in comparing models. We will use this at first to compare to the “block-model” which only includes the constant. When interpreting the estimates from a logistic regression, we must remember that the b values are changes in logits…the expected change in logit units for a one unit increase in the predictor variable. Exponentiating the b value gives us the expected change in odds (p/p-1) for a one unit increase in X. We can use this with a multiplicative interpretation. If our odds ratio is less than 1, then we can either mirror the variable and rerun or compute the inverse and use the phrase “times greater” with a 1 unit decrease in X value. However, the percentage interpretations can remain (Odds are decreased by X%). How do we predict, now? We plug and play but we need to remember our terms (especially in solving for p if we have our intercept and b1 and X). We can then see how different levels of X change the value of Y and see if it got it in the right category (based on a cut value of probability—usually .5--). the P will be the distribution of the odds…basically the probability that Y is 0 or 1. Specificity is ability to reject the negatives. Sensitivity is ability to detect the positives. The number correct over total number would be the classification rate… If we want to use more variables to predict Y, then we wants to use a likelihood ratio test to compare deviance values to see if adding new is better. The deviance change id distributed on chi square. Similar to comparing R2 change. We can also add interactions. There is also a probit alternative where we look at what z value cuts of a particular percentage to the left. The logit coeffecients and standard errors are 1.7x larger than the probit. This answers the question…”for a given proportion…what z value cuts off that percentage to the left?”. If including new variable, you must meet the proportion odds assumption. Always interpret coefficients in terms of log odds of a higher category…predicting the probability of being in a particular category relative to a reference category (if dummy coded).