Introduction to R: kNN classification
advertisement

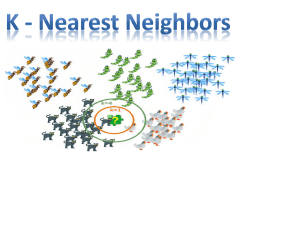
Classification by nearest neighbor strategy.
Lazy Learning via kNN algorithm. Nearest Neighbor Algorithm.
Given a dataset of cancer biopsies, each with several features, we
want to train a classifier so that it can figure out whether a
specific sample(s) represent a benign or cancerous case.
Classification via nearest neighbor approach has following
characteristics.
Pros: (a) simple and effective, (b) makes no assumption of any
distribution, and (c) fast training phase
Cons: (a) Does not produce any model (no abstraction, hence
Lazy Learning), (b) classification process is slow, (c) needs a lot
of memory, (d) nominal features and missing data need
additional scrutiny.
Training dataset: Examples of classification into several
categories.
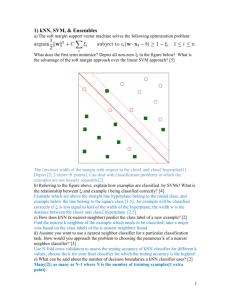
For each record in the test dataset, kNN identifies 𝑘 known
samples nearest in similarities. Unlabeled test is assigned the
category label of the majority of the k nearest neighbors.
𝑘 = 5 kNN environment
The distance function between the target object and an accepted
vector sample (the ‘geographical’ distance) could be measured in
many ways. By default, though, it is Euclidean distance metric.
Distance between (𝑥1 , 𝑦1 ) and (𝑥2 , 𝑦2 ):
Euclidean: √(𝑥1 − 𝑥2 )2 + (𝑦1 − 𝑦2 )2
Manhattan: |𝑥1 − 𝑥2 | + |𝑦1 − 𝑦2 |
Minkowski: (|𝑥1 − 𝑥2 |𝑞 + |𝑦1 − 𝑦2 |𝑞 )1/𝑞
We are going to use Wisconsin breast-cancer data available in
our website. R uses Euclidean distance metric.
Major issue in kNN classification approach is the value of 𝑘. If 𝑘 is
large, one is basically focusing on majority decision, regardless of
which clusters are nearest to it.
If 𝑘 is too small, noisy data or outliers may control the situation.
This is not acceptable either. We need 𝑘 that avoids two
extremes.
Cluster separation boundary
Low k.
For any k, we could encounter misclassifications: False Positive,
and False Negatives. 𝑘 needs to be chosen so that these
misclassifications tend to be as low as possible.
First we read the data as a csv file from our website, and figure
out its structure.
> md=read.csv("http://web.cs.sunyit.edu/~sengupta/num_maths/wisc_bc_data.csv").
Now let’s look at its structure. We use str() operation to reveal
the data structure md.
> str(md)
It seems there are 32 variables (10 × 3 + 2) over 569 samples.
ID we do not need. The Diagnosis is our focus, and it is a factor
with 2 levels: B (Benign), M (Malignant). Note that all features
are numeric.
We now filter the data, and need to prepare it further.
> table(md$diagnosis)
B M
357 212
Majority is Benign. Let us use proportion on the diagnosis factor
to find what proportion leads to benign.
> > round(prop.table(table(md$diagnosis))*100, digits=1)
B M
62.7 37.3
We are going to focus on three features of our data. You have to
try all of them or some other combination of features.
>summary(md[c("radius_mean","area_mean","smoothness_mean")])
radius_mean
Min. : 6.981
1st Qu.:11.700
Median :13.370
Mean :14.127
3rd Qu.:15.780
Max. :28.110
>
area_mean
Min. : 143.5
1st Qu.: 420.3
Median : 551.1
Mean : 654.9
3rd Qu.: 782.7
Max. :2501.0
smoothness_mean
Min. :0.05263
1st Qu.:0.08637
Median :0.09587
Mean :0.09636
3rd Qu.:0.10530
Max. :0.16340
Everything is okay, except our data need to be polished further.
kNN is very sensitive to measurement scale of the input features.
In order to make sure our classifier is not comparing apples with
oranges, we are going to rescale each of our features (except the
diagnosis, that is a factor) with a function
> normalize <- function(x) {
+ return((x-min(x))/(max(x)-min(x)))
+}
>
>
>
>
# lets try on a vector
q=c(12,56,3,2.7, 9.8,12.6,-5.3)
q=normalize(q)
q
[1] 0.2822186 1.0000000 0.1353997 0.1305057 0.2463295 0.2920065
0.0000000
The minimum and the maximum are at 0.0 and at 1.0.
We are going to normalize these numerical data, and make a data
frame with the normalized attributes.
To apply to our data, we are going to call lapply on column 2:31
using normalize function.
> #redefine our data frame md by normalizing first all the numerical
> # columns in our dataframe. We might need them later.
> #apply normalize function to all our data frame columns. We need to
apply lapply.
> md=as.data.frame(lapply(md[,2:31], normalize))
Anytime you want to create a new data frame that might contain
mixed data you need to make the call as.data.frame.
Let’s pick the three attributes of our interest and put them in a
new data frame qq.
qq=data.frame(md$area_mean,md$radius_mean,md$smoothness_mean)
> summary(qq)
Check every attribute is bounded between 0 and 1.
We are now ready to define our training set and our testing set.
We are going to split the data into two portions: mdn_train, and
mdn_test. The first 469 rows (samples) would comprise the
training set, and the rest 100 rows the mdn_test
>
>
>
>
train=qq[1:469,] # First 469 rows for training
test=qq[470:569,] # Last 100 rows for our testbed
train_labels=q1[1:469,1] #Our training classification factors
test_labels=q1[470:569] # Our target sample factors
The classifier would change test_labels column if necessary.
The kNN package is in “class” that we need to install. You may need to
make
install.packages(“class”) first from a mirror site. And then call
library(class)
> library(class)
> p=knn(train, test, cl=train_labels, k=3)
>
Thank Heaven. It worked. Now, we need to do some tidying it
up. For this we need a package called “gmodels” , which will
provide us with cross tabulation results.
> library("gmodels")
> CrossTable(x=test_labels, y=p, prop.chisq=FALSE)
The final output emerges now. We don’t want Chi square results.
There were 100 test data. The top-left data (True Negative)
score is 56/61 (we suggested 61 of them “benign”, the classifier
picked 56 of them benign). The top-right data is False Positive,
meaning 5 of the 61 were misclassified under our scheme. The
bottom left box implies total number of False Negatives, 10 out of
39 were wrongly classified as Malignant. The bottom right box
shows True Positive stats: 29 out of 39 were correctly labelled.
Perhaps we would get different result using different k values.
Find out how our misclassification results change by selecting
different k values. We try with k=7
> p=knn(train, test, cl=train_labels,k=7)
> CrossTable(x=test_labels, y=p, prop.chisq=FALSE)
Now we are making it slightly worse. Look at the cross tabulation
figures.
The top row didn’t change, but now we made 1 more
misclassification in the False Negative area. Now you know how
you have to approach the problem.