tpj12757-sup-0015-Legends
advertisement

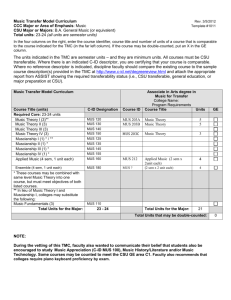
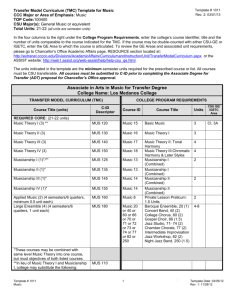
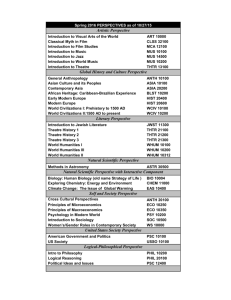
SUPPORTING INFORMATION LEGENDS SUPPORTING FIGURE S1. The MUS open reading frame is 3,586 base pairs long and lacks introns. (a) Schematic diagram of the MUS gene (At1g75640) showing locations of the mus alleles. The two alleles that harbor point mutations are indicated with vertical black lines. These mutations result in a G (glycine) to R (arginine) transition in mus-1, and a W (tryptophan) to stop transition in mus-2. The location of the T-DNA insertion is marked by a white triangle. (b) Relative mRNA levels in the three mus alleles. SUPPORTING FIGURE S2. mus does not disrupt organelle distribution. (a-f) Red fluorescence corresponds to Propidium Iodide (PI) staining of cell walls. Scale bars = 5 µm. (a-c) Green fluorescence represents the vacuolar membrane marker 35Spro:delta-TIP:GFP (Cutler et al., 1999). Both WT (a) and mus (b-c) stomata contain several vacuoles. (d-f) Green fluorescence corresponds to 35Spro:TUA:GFP localization. Each micrograph depicts the mid-plane of a stoma, where 35Spro:TUA:GFP marks perinuclear MTs surrounding the nucleus. Nuclei in both WT (d) and mus (e-f) stomata are centrally located near the ventral wall. Chloroplasts (red autofluorescence) are also visible along both the dorsal and ventral walls. 1 SUPPORTING FIGURE S3. Association between the direction of MT growth and the degree of symmetry disruption in mus-1 stomata. Cell walls visualized utilizing Propidium iodide (PI) staining. Images were converted to gray scale for clarity. Y-axis represents direction of MT growth (expressed as the number of inbound (blue) and outbound (red) comets). The Xaxis indicates the degree of disruption of morphological symmetry in stomata of different backgrounds. On the X-axis the phenotypic severity increases from left to right, with the left most micrograph representing a normal (wild-type) stoma and the right most micrograph representing the most severely disrupted stomatal phenotypes observed in mus-1. Note that higher levels of symmetry disruption are associated with an increase in the number of inbound comets (blue squares), and a decrease in the number of outbound comets (red triangles). SUPPORTING FIGURE S4. MUS maintains bilateral symmetry via acting after symmetric division but before pore formation. (a, c, d, f) Red fluorescence corresponds to cell walls visualized by PI staining. (b, e) Green fluorescence of microtubules derives from 35Spro:TUA:GFP expression. Scale bars = 5 µm. (a) Recently divided WT stomata prior to pore formation. Formation of a new cell wall normally generates two symmetrical young GCs. The young newly formed wall is visualized through propidium iodide. (d) Recently divided mus-1 stomata prior to pore formation. As in WT, the new wall in young mus-1 stomata form two symmetrically arranged GCs. (b-c) Radial MT arrays in young WT stomata are symmetrically arranged around the future pore site. (c) 2 Micrograph of b, with identical levels of intensity and fluorescence, but with green channel removed in order to make the young GC wall visible. (e) MT arrays are not symmetrically arranged around the future pore site in mus, and instead appear to concentrate at the center of each GC. (f) Micrograph of (e), with identical levels of intensity and fluorescence, but with green channel removed. All scale bars = 5 µm. SUPPORTING FIGURE S5. MUS orthologs are present in diverse plant lineages. An unrooted, maximum likelihood phylogeny shows that MUS orthologs are present in plant lineages ranging from mosses and liverworts to dicots and monocots. Each ortholog’s gene ID, or clone name, is contained in brackets adjacent to the corresponding ortholog. SUPPORTING FIGURE S6. MUS localization patterns remain consistent throughout multiple plant organs. (a-j) Red fluorescence corresponds to cell walls visualized by PI staining (except in (h). Green fluorescence represents MUSpro:MUS:tripleGFP localization. (a-c, f-g) 3 Scale bars = 15 µm. (d, f, i) Scale bars = 30µm. (a-b) Stem. (c) Sepal. (d-e) Petal. (f-h) Anther. (h) Micrograph of (g), with identical levels of intensity and fluorescence, but with red channel removed. (i) Stigma (j) Style. Arrowheads signify GMCs, while asterisks signify mature stomata. SUPPORTING FIGURE S7. MUS localizes to the cell membrane. Red fluorescence corresponds to FM4-64 cell membrane (PM) staining; green fluorescence derives from MUSpro:MUS:tripleGFP expression. (a) Meristemoid (left) and a GMC (lower right) shown in face view (paradermal optical section) that resulted from two asymmetric divisions. At the left, MUS expression is localized in the new cell wall of the meristemoid, but not in the newly formed pore wall in the GMC (lower right). The brightness in all micrographs was increased to demonstrate overlaps in localization. (b-c) Both micrographs depict different planes of section of the same newly divided GMC shown in (a). White dashes in (b) indicate where the plane of section was obtained. In (c), the GMC is viewed in an optical section perpendicular to the plane of the epidermis in (b). Yellow fluorescence indicates overlap between FM4-64 stain and MUSpro:MUS:tripleGFP localization. (d-e) Both micrographs depict different planes of section of the same meristemoid shown in micrograph (a) (at the left). White dashes in (d) indicate the location that the plane of section e was obtained from. In (e), the meristemoid is viewed in a plane of section that is anticlinal to the plane of the epidermis in (b). Yellow 4 fluorescence indicates the overlap between FM-464 & MUSpro:MUS:tripleGFP localization (a). Scale bar = 10 µm and b. Scale bar = 5µm. SUPPORTING FIGURE S8. MUS does not localize to the cortical Endoplasmic Reticulum. Green fluorescence represents MUSpro:MUS:tripleGFP, while red fluorescence corresponds to Hexyl Rhodamine staining of the cortical Endoplasmic Reticulum (ER). The first column of micrographs represents both MUS and ER signal, the second column corresponds to only MUS signal, and the third column represents only ER signal. Scale bars = 5 µm. (a) Meristemoid. (b) Guard Mother Cell. (c) Mature Stoma. SUPPORTING FIGURE S9. mus displays additive phenotypes with several other mutants known to disrupt stomatal development. (a) Wild type Col-0 gl1 stoma exhibiting normal bilateral symmetry. (b) mus-1 stoma displaying disrupted bilateral symmetry and morphogenesis. (c) Stomatal cluster in flp-1 with two bilaterally symmetrical stomata. (d) Stomatal cluster in a flp-1 mus-1 double mutant containing one stoma (left) displaying normal bilateral symmetry, and another stoma (right) with disrupted symmetry. (e) A tumour like cell cluster in a fama-1 single mutant (which entirely lacks guard cells). (f) A tumour-like cluster in a fama-1 mus-1 double mutant (that displays no obvious stomatal skewing) (g) Stomatal cluster in tmm-1 single mutant containing bilaterally symmetrical stomata. (h) Cluster in a tmm-1 mus-1 double mutant 5 containing two bilaterally symmetrical stomata (left), and two with disrupted symmetry (right). (i) Single Guard Cell (SGC) in 35S:CDKB1;1 N161 overexpression mutant line. (j) 35S:CDKB1;1 N161 mus-1 double mutants contain SGCs (left) as well as stomata with disrupted bilateral symmetry (right). All scale bars = 5 µm. SUPPORTING FIGURE S10. Comparison of conserved amino acid motifs indicates that MUS likely encodes an atypical kinase (modified from Castells and Casacuberta, 2007). Comparison of conserved amino acid motifs (red box) in Kinase VII domains of typical (DFG) and atypical receptor-like kinases (adapted from Castells & Cascuberta, 2007). All proteins shown are in Arabidopsis except for MARK. MUS: MUSTACHES; MARK: MAIZE ATYPICAL RECEPTOR KINASE; TMKL1: TRANSMEMBRANE KINASE LIKE1; SUB: STRUBBELIG; CRR1: CRINKLY4RELATED1; CRR2: CRINKLY4-RELATED2 BRI1:BRASSINOSTEROID INSENSITIVE1. Note that in the wild-type MUS protein, a glutamic acid (E) is substituted for aspartic acid (D). The presence of this substitution suggests that MUS might be incapable of phosphorylation. SUPPORTING TABLE S1. Nature and location of mutations carried in three mus alleles. 6 SUPPORTING MOVIE S1. 35Spro:EB1:EB1: GFP movement in a WT stoma. SUPPORTING MOVIE S2. 35Spro:EB1:EB1: GFP movement in a skewed mus-1 stoma. SUPPORTING MOVIE S3. 35Spro:EB1:EB1: GFP movement in an abnormally expanded mus-1 stoma. 7