Discrete Joint Distributions
advertisement

BIOINF 2118
Estimation Part 4
Page 1 of 5
The Student’s t confidence interval; pivotal quantities.
Here we will take into account the fact that in some cases we really don’t know the variance.
It has to be estimated.
Example:
Data about cheese:
X=c(0.86, 1.53, 1.57, 1.81, 0.99, 1.09, 1.29, 1.78, 1.29, 1.58).
(measurements of acid concentration in cheese).
Goal: 90% confidence interval.
Notation: in the book,
,
Then
,
.
.
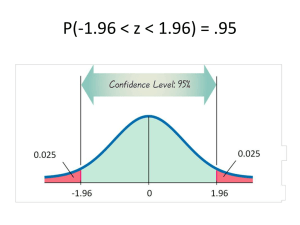
The approximate interval we got before is
=
= mean(x)+c(-1,1)*qnorm(0.95)*sd(x)/sqrt(n)
= (1.208565, 1.549435).
Suppose that in reality
,
and we use the same “recipe” to construct confidence intervals.
What is the real coverage probability? See meanCoverageOfConfidenceIntervals.R
Answer: around 86%, lower than the 90% claimed.
Doesn’t sound bad, but, the NON-coverage is 14% instead of 10%, which is 40% worse.
The reason it’s off: our “recipe” assumes we know the true sd(x), but we don’t.
So let’s back up & try again. First, a detour.
The Chi-Square distribution
If
Z1,...,Zn ~ Norm(0,1) i.i.d., then the sum of squares is a chi-square random variable:
~ “chi-square on n degrees of freedom”.
This the identical distribution as Gamma(n/2, 1/2), i.e. with shape parameter n/2 and rate parameter
½ (or scale parameter 2). For example, for n = 2, the squared distance
point formed by i.i.d. standard normals is Exponential with scale parameter 2.
from the origin of a
BIOINF 2118
Estimation Part 4
Page 2 of 5
The joint distribution of the sample mean and sample variance: i.i.d. NORMAL.
The key fact:
for the i.i.d normal case,
,
a)
(the degrees of freedom are reduced by 1 because the mean is estimated).
b) X and
are STATISTICALLY INDEPENDENT.
The quantity
is unknown because the true variance is unknown.
pchisq(K,n – 1).
However, we can say for sure that
We can turn that into a probability statement focusing on
(though
is NOT the r.v.).
pchisq(K,n – 1).
Therefore, if we want a 90% confidence interval for , for example, we set the probability (right-hand
side of the equation) to 0.05 and then to 0.95, and remember that pchisq and qchisq are inverse
functions, so that K becomes (respectively)
qchisq(0.05,n – 1) and qchisq(0.95,n – 1).
Example: cheese data.
For n=10, qchisq(0.05,n – 1) = 3.325 and qchisq(0.95,n – 1)=16.92.
(Notice the lost degree of freedom!)
and
In our data, the sample standard deviation is
.
.
So the endpoints are
The UPPER point 0.2906 corresponds to the LOWER probability (0.05).
(And the LOWER point 0.0571 corresponds to the UPPER probability (0.95).)
Why? because the upper point is the value at which the data are at the LOW end of possible
outcomes.
BIOINF 2118
Estimation Part 4
Page 3 of 5
The significance test interpretation of confidence intervals
The confidence interval endpoints are the places where the data are at the boundary of plausible
probability.
Lower
CI point
observed
Upper tail
Upper
CI point
Lower tail
The t distribution
The t will help us provide an “honest” CI for the normal mean.
If
where Z is standard normal and Y is chi-square on
then T is distributed as a random variable.
degrees of freedom, and
,
Notation: independent
Properties:
As
, this approaches the standard normal.
For small , it has fat tails.
For
, they are so fat that the mean and variance don’t exist (Cauchy distribution).
BIOINF 2118
Estimation Part 4
Page 4 of 5
Because of the key fact (a) and (b) above, define
and
.
Then
But this involves only
.
. So we can now construct an EXACT confidence interval for
NOT require the “plug-in principle”, pretending that we know
.
that does
BIOINF 2118
Estimation Part 4
Page 5 of 5
Cheese example; confidence intervals (“meanCoverageOfConfidenceIntervals.R”.):
### Based on the normal approximation
> mean(x)+c(-1,1)*qnorm(0.95)*sd(x)/sqrt(n)
[1] 1.208565 1.549435
### Based on the t distribution
> mean(x)+c(-1,1)*qt(0.95,9)*sd(x)/sqrt(n)
[1] 1.189058 1.568942
## from "11-estimation Part 4.doc"
cheeseX=c(0.86, 1.53, 1.57, 1.81, 0.99, 1.09, 1.29, 1.78, 1.29, 1.58)
simulateConfidenceInterval = function(X=cheeseX, CImethod=c("normal", "t"),
simMethod=c("normal","Bootstrap"), nsims=10000, nreps=2) {
CImethod = CImethod[1]; simMethod = simMethod[1]
mu = mean(X); sig = sd(X); n = length(X)
simFunction = function(repNumber) {
x = switch(simMethod,
normal=rnorm(n, mu, sig),
Bootstrap=sample(X, 10, replace=TRUE) )
interval = switch(CImethod,
normal=mean(x)+
c(-1,1)*qnorm(0.95)*sd(x)/sqrt(n),
t =mean(x)+
c(-1,1)*qt(0.95,9)*sd(x)/sqrt(n)
)
covers = (mu > interval[1]) & (mu < interval[2])
return(covers) ## Boolean
}
unix.time( {
for(rep in 1:nreps) {
coverResult = sapply(1:nsims, simFunction)
cat(mean(coverResult), " CImethod=", CImethod, " simMethod=", simMethod, "\n")
}
})
}
simulateConfidenceInterval()
simulateConfidenceInterval(CImethod="t")
simulateConfidenceInterval(simMethod="Bootstrap")
simulateConfidenceInterval(CImethod="t", simMethod="Bootstrap")