jec12111-sup-0001-TableS1-S2-FigS3
advertisement
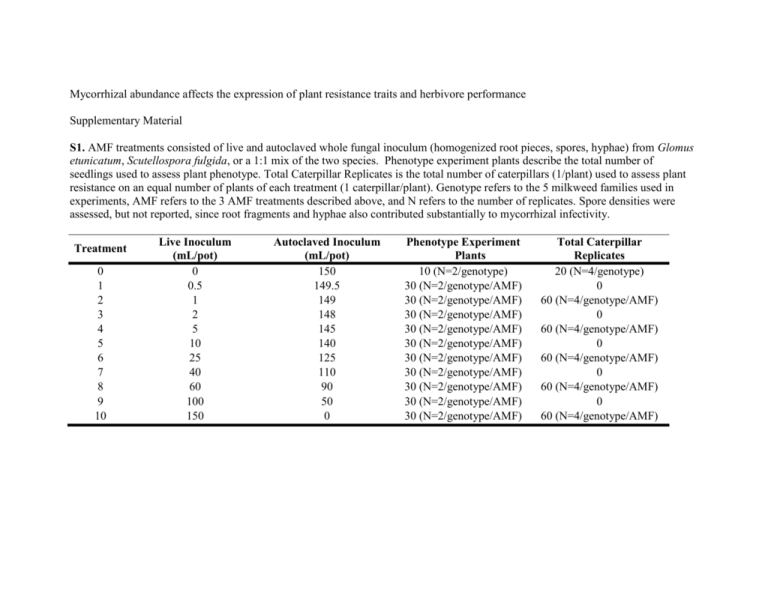
Mycorrhizal abundance affects the expression of plant resistance traits and herbivore performance Supplementary Material S1. AMF treatments consisted of live and autoclaved whole fungal inoculum (homogenized root pieces, spores, hyphae) from Glomus etunicatum, Scutellospora fulgida, or a 1:1 mix of the two species. Phenotype experiment plants describe the total number of seedlings used to assess plant phenotype. Total Caterpillar Replicates is the total number of caterpillars (1/plant) used to assess plant resistance on an equal number of plants of each treatment (1 caterpillar/plant). Genotype refers to the 5 milkweed families used in experiments, AMF refers to the 3 AMF treatments described above, and N refers to the number of replicates. Spore densities were assessed, but not reported, since root fragments and hyphae also contributed substantially to mycorrhizal infectivity. Treatment 0 1 2 3 4 5 6 7 8 9 10 Live Inoculum (mL/pot) 0 0.5 1 2 5 10 25 40 60 100 150 Autoclaved Inoculum (mL/pot) 150 149.5 149 148 145 140 125 110 90 50 0 Phenotype Experiment Plants 10 (N=2/genotype) 30 (N=2/genotype/AMF) 30 (N=2/genotype/AMF) 30 (N=2/genotype/AMF) 30 (N=2/genotype/AMF) 30 (N=2/genotype/AMF) 30 (N=2/genotype/AMF) 30 (N=2/genotype/AMF) 30 (N=2/genotype/AMF) 30 (N=2/genotype/AMF) 30 (N=2/genotype/AMF) Total Caterpillar Replicates 20 (N=4/genotype) 0 60 (N=4/genotype/AMF) 0 60 (N=4/genotype/AMF) 0 60 (N=4/genotype/AMF) 0 60 (N=4/genotype/AMF) 0 60 (N=4/genotype/AMF) S2. AICc values comparing linear, saturating, and logistic models to root colonization of Asclepias syriaca by arbuscular mycorrhizal fungi Glomus etunicatum, Scutellospora fulgida, and a mix of the two species. Inoculum densities follow those given in Supplementary Table 1. Null Linear Saturating Logistic GE -42.0 -89.2 -110.4 -87.3 SF -24.6 -69.7 -71.9 -64.0 Mix -94.9 -99.2 -132.4 -102.3 0.2 0.2 0.4 0.6 0.8 0.0 R2 = 0.146 0.1 0.2 0.3 0.4 0.5 0.6 0.1 0.2 0.3 0.4 0.5 0.6 0.1 0.2 0.3 0.4 0.5 0.6 0.5 0.6 0.1 0.3 0.3 f) 0.1 0.0 g) h) 0.2 0.4 0.6 0.8 0.0 R2 = 0.0422 i) 0 0.0 2 2.0 4 0.0 0.1 0.2 0.3 0.4 0.5 0.6 0.7 0.0 R2 = 0.0919 k) j) 0.2 0.4 0.6 0.8 R2 = 0.275 0.0 0.1 0.2 0.3 0.4 0.5 0.6 0.7 0.0 p) q) 0.4 0.6 0.8 R2 = 0.0677 0.2 0.4 0.0 0.0 1.5 3.0 n) 0.2 0.6 R2 = 0.0748 0.2 0.3 0.4 0.1 0.2 0.3 0.4 0.5 R2 = 0.0283 r) 2 2 6 5 0.1 o) 0.0 10 m) G. etunicatum R2 = 0.0659 l) 0.005 0.0 0.0 1.5 3.0 0.0 0.1 0.2 0.3 0.4 0.5 0.6 0.7 0.0 0.1 0.2 0.3 0.4 0.5 0.6 0.7 0.0 0.010 0.0 0.1 0.2 0.3 0.4 0.5 0.6 0.7 8 0.0 1.5 3.0 0.8 0.6 e) 6 Foliar Cardenolides R2 = 0.0376 c) 0.2 0.8 0.2 d) 0.020 %P 0.0 0.10 0.30 0.0 1.5 3.0 Latex 0.0 0.1 0.2 0.3 0.4 0.5 0.6 0.7 2 Foliar Trichomes R2 = 0.0125 b) 0.000 SLM a) 0.020 AG Biomass S3. Best-fit lines relating the degree of AMF colonization of Glomus etunicatum, Scutellospora fulgida, or a mix of the two species to the expression of plant nutrition and resistance traits in Asclepias syriaca plants. Each point represents a plant individual. Traits include a-c) aboveground (AG) biomass, d-f) % foliar phosphorus, g-i) latex exudation (mg), j-l) specific leaf mass (SLM), m-o) total foliar cardenolides, and p-r) the density of foliar trichomes on the bottom surface of the leaves. 0.0 0.2 0.4 0.6 0.8 S. fulgida Proportion of Plant Root Colonized by AMF 0.0 0.1 0.2 0.3 Mix 0.4 0.5 0.6 S4. Proportion of total variance in the expression of Asclepias syriaca nutrient and resistance traits explained by AMF abundance, AMF species identity, plant genotype, the presence or absence of AMF, and interactions between genotype (G) and all other AMF predictors, or the variance remaining (error). Numbers indicate the proportion of variance explained by each significant predictor (p<0.05), calculated by dividing the sums of squares explained by each factor by the total variance in each trait, among all phenotype assay plants (a-f). a ) AG Biomass b)%P error c ) Latex error G X Presence 0* G X Presence G x Abundance 0* G x Abundance error 0.02 * 0.03 * G X Species 0.02 * G X Species 0* G X Species 0* AMF Presence 0* AMF Presence Plant Genotype 0.01 * Plant Genotype 0* Plant Genotype 0.93 * AMF Species 0.2 0.4 0.6 0.8 1.0 1.2 0.0 d ) SLM 0.2 0.4 0.6 0.8 0* 0.01 * G X Presence 0.02 * G x Abundance 0.02 * G X Species 0.02 * AMF Presence 0.03 * 0.04 * Plant Genotype 0.0 0.2 0.02 * 0.01 * 0.6 0.8 1.0 0.0 0.2 0.17 * 0.4 0.51 * AMF Species 0.55 * AMF Abundance 0.4 0.8 0.06 * Plant Genotype 0.35 * AMF Species 0.16 * AMF Abundance 0* G X Species Plant Genotype 0.6 f ) Trichome Density 0.01 * AMF Presence 0.4 error G X Presence 0* 0.02 * 0.2 G x Abundance AMF Presence AMF Species 0.0 e ) Foliar Cardenolides 0.02 * G X Presence 0.46 * 0.04 * AMF Abundance error G x Abundance 0.3 * 0.1 * AMF Species 0.12 * AMF Abundance error G X Species 0.44 * AMF Species 0.04 * 0.0 0.05 * G x Abundance AMF Presence AMF Abundance 0* G X Presence 0.01 * 0.6 0.18 * AMF Abundance 0.8 Proportion of Variance Explained 0.0 0.2 0.4 0.6 0.8








