mec12219-sup-0001-TableS1
advertisement

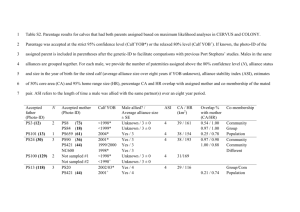
Supporting Information Microsatellite genotyping For the DNA extraction of blood samples we used the DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany) following the customers instructions. Polymerase chain reaction (PCR) was performed in a 10µl volume consisting of 1x QIAGEN PCR buffer (containing TrisCL, KCL and (NH4)2SO4), 1.5 mM MgCl2, 0.3 mM of dNTP, 0.5 µl of each of 10 µM forward and reverse Primers, 0.5 U HotStar Taq Polymerase (Qiagen) and 1 µl template. DNA amplification was then performed in an Eppendorf Mastercycler Gradient (Eppendorf GmbH, Hamburg, Germany) starting with an initial denaturation step at 95°C for 5 min. The cycling parameters were: 20 cycles at 94°C for 30 sec, an annealing temperature of 62°C (decreasing 0.5°C per cycle) followed by an elongation step at 72°C for 30 sec, followed by 15 cycles at 94°C for 30 sec, 52°C for 30 sec and 72°C for 45 sec. Finally an elongation step at 72°C for 40 min. The annealing temperature was held fixed at 58.3°C for the primer PyrOr6. All forward primers (5´-end) were labeled with the fluorescent dyes 6-FAM or HEX. Genotyping of individuals was performed with an ABI Sequencer 3130xl (Applied Biosystems, HITACHI, Foster City, USA) using the software GeneMapper ID-X 1.1 (Applied Biosystems). We were able to genotype 99% of the 18 loci for n = 163 individuals. Deviation from Hardy-Weinberg equilibrium and null allele frequencies were calculated with CERVUS 3.0 (Kalinowski et al. 2007). The null allele frequency of our microsatellite marker set ranged from -0.07 to 0.08 (table 1; main text); none of the microsatellite markers deviated from Hardy-Weinberg-equilibrium. Linkage disequilibrium was tested between all pairs of loci with the software FSTAT 2.9 (Goudet 2001), which uses a Bonferroni correction of significance test for multiple pairwise comparisons. We tested linkage for all adult individuals (n = 76) captured within the study area. Furthermore, linkage was tested separately for all individuals sampled (adults and their offspring breeding in nest boxes) in each breeding season. Linkage disequilibrium was detected when testing the data set of adult individuals (Cyanp5xPyrOr4, PyrOr11xCyanp8, PyrOr2xUnaCT43), but did not emerge consistently within the data sets of each breeding season. Parentage analysis The computer software CERVUS 3.0 (Kalinowski et al. 2007) estimates parentage based on a likelihood approach. To estimate the likelihood of an individual being the true parent of a given juvenile, a simulation is implemented accounting for the number and proportion of candidate parents sampled as well as for potential genotyping errors. The average non-exclusion probability of our microsatellites, the probability that an individual is erroneously assigned as true parent, was calculated using the whole data set of n = 163 individuals. The average non-exclusion probability of the 18 loci used was 0.003% for the first parent. The mean proportion of individuals typed was 99% and typing error was set to 0.01 for all analysis. We first conducted a within-group parentage assignment. To account for individuals of a given group potentially not sampled, we set the number of candidate parents sampled to the maximum number of same sex adult individuals per group and year plus one (see also Richardson et al. 2001), and we set the proportion of sampled parent individuals to 85% for each breeding season, respectively. Parentage was then assigned separately for females and males and each breeding season (2008/2009: nchicks = 31, nadult female = 13, nadult male = 23; 2009/2010: nchicks = 39, nadult female = 18, nadult male = 26; 2010/2011: nchicks = 34, nadult female = 20, nadult male = 24). When a female was assigned with no more than two mismatches (between her own and the offspring genotype) and 95% confidence or no more than one mismatch and 80% confidence, she was considered the true mother. In a second step, paternity was assigned based on the 1 known mother following the same procedure as for females. Furthermore, assigned female-male pairs were only regarded as true parents if the trio confidence, the likelihood that the male-female-pair are the true parents, was >80%. The whole procedure was then repeated on the pool of all adult individuals (sampled until a given breeding season) as potential parents (2008/2009: npot. mother = 17, npot. father = 29; 2009/2010: npot. mother = 37, npot. father = 43; 2010/2011: npot. mother = 59, npot. father = 69). This was done to account for possible extra-group parentage. The proportion of sampled parents was specified as described above. Marshall et al. (1998) already noted that first-order kin of the true parent pair can have a higher LOD (log likelihood ratio) score than a parent individual and will therefore erroneously be considered as true parent. When parentage was unclear, we followed two different procedures. First, we compared trio LOD scores of the two most likely parent pairs and assigned those with the highest LOD score as true parents. Additionally, we conducted a parent pair assignment where both individuals are assigned simultaneously. We used the same settings (typing error and proportion sampled father/mother) as described for the within-group assignment. Table S1 Number of pairs (single breeders) and groups (pairs with helpers) captured in Buenaventura in three consecutive seasons. The mean group size comprises only pairs with helpers. The number of lost broods is summarized for pairs and groups. The number of second brood events (after brood loss) is specified in brackets. For all numerical specifications, only those breeding events were included where the whole group could be captured before brood loss. Season Number of pairs Number of groups Mean group size Number of broods lost 2008/2009 6 7 3.7 7 (3) 2009/2010 2 10 5.5 2 (0) 2010/2011 1 9 5.3 1 (0) 2