Willoughby supplementary text 081215
advertisement

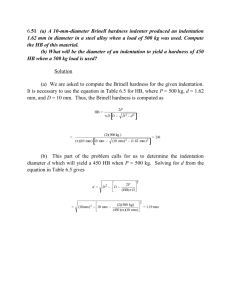
Supplementary Material and Figures ad ‘A scalable label-free approach to separate human pluripotent cells from differentiated derivatives’ by NA Willoughby et al. Atomic Force Microscopy: data acquisition. 5 All AFM work was carried out using the NanoWizard III Bio AFM (JPK Systems, Germany), mounted on a Zeiss Observer D1 inverted optical microscope (Carl Zeiss Group, Germany) equipped with phase contrast and fluorescence imaging. The AFM setup was placed on a Halcyonics i4 anti-vibration table (Accurion, Germany) and acoustically isolated to reduce the influence of background vibration and noise. For all measurements Pyrex-Nitride probes (PNP- 10 DB, NanoWorld, Switzerland) were used which had spring constants between 0.051 and 0.063 N/m (determined by the thermal fluctuations method). Tips were pyramidal with a macroscopic half-cone angle of 35º and a height of 3.5µm. The tip curvature was typically less than 10nm. Once mounted under the AFM, optical microscope imaging was used to position the probe over the region selected for measurement (Fig. 1A). These regions were chosen to represent the 15 dominating characteristics of the samples. For each sample several regions were measured where each of these measurements consisted of hundreds of individual force indentation curves. The individual force indentation curves were distributed over the sample regions in the form of a rectangular grid such as the one shown in figure 1A. In wells containing confluent cell populations these regions covered several cells, while in sparsely populated wells also individual 20 cells were measured. For the transgenic cell line H1-Oct4-EGFP individual pluripotent cells could easily be identified by fluorescence, while phase contrast imaging was used to identify suitable cells of the other lines. Willoughby et al Suppl 08DEC2015 1 Atomic Force Microscopy: data analysis Representative cell stiffness values were obtained by measuring several thousand force versus indentation curves for each cell type using AFM. These curves were subsequently analysed using 5 an automatic computational analysis algorithm developed by ourselves. To extract information about the stiffness of the cells only the unmodified approach curves were used, that is, the restoring force exerted by the cells on the AFM tip as a function of the indentation depth. The most challenging aspect of the algorithm is the identification and exclusion of force versus indentation curves that carry some measurement error. Since we were investigating cells, i.e., 10 very soft material, we refrained from correcting any baseline tilt, but did discard all curves that had a baseline tilt larger than 50 µN/m. All curves with “jumps and bumps” in the region relevant for the fitting were also discarded. Although a jump or bump in a force versus indentation curve is not necessarily a measurement error and could very well represent an actual response of the cell, it is inconsistent with the indentation model we were using to obtain the 15 elastic modulus and would therefore lead to meaning-less results. This fastidious procedure automatically identified nearly all curves that showed some measurement error, which could then be discarded. Analysis of the remaining force versus indentation curves began with removing any force offset. Initially this is done using the baseline far away from the estimated contact point. Later, once the 20 contact point was precisely located, the baseline near the contact point was used. The contact point is initially located by pricewise fitting of a straight line starting at the deepest indentation moving upward until the first derivative of the line approaches zero. Subsequently, the Hertz model (for a conical indenter) Willoughby et al Suppl 08DEC2015 2 f (d) = E 2 tan(a ) 2 d (1- n 2 )p was fitted to the force versus indentation curve several times, where f is the force, d the indentation depth, E Young’s modulus, a the half cone angle and n the Poisson ratio, for 5 which we use the customary value of 0.5. Following each fit, the contact point was updated and the fitting range adjusted. All results presented in this manuscript have been obtained by fitting from 300nm below the contact point to 50nm above it. Only results from high quality fits were included in the data analysis, while all force curves that resulted in a fitting error (essentially the square root of the mean squared residual of the least squares fit) of more than 0.04nN were 10 excluded. From the fit a Young’s modulus, which we dubbed Cell Elastic Modulus (CEM), was obtained as a measure of the local stiffness of the cells. For each cell type all results were then combined and analysed. Initially a probability distribution of CEMs was determined by ordering the measured elasticities into bins of equal width of 0.5kPa (see Figs 1B and S1). From these distributions the locations of the probability maxima were 15 derived. The median and mean were obtained directly from the cumulative distributions of the unbinned values (Figures 1B and S1). All confluent samples showed a smooth distribution with a maximum in the kPa region, while some non-confluent samples showed a second, well separated maximum in the MPa region. As this second part stems from the plastic material on which the cells were cultured, force versus indentation curves with CEMs of more than 0.1MPa were 20 excluded. The resulting distributions often showed a very thin but long “tail”. These measurements are likely non-representative, but would have a very large effect on the mean and median. To eliminate this effect, the probability distributions were cut off at a CEM value beyond which all bins had fewer than 10 counts (less than 2 for H1-hESC p65, H1-hESC p68, Willoughby et al Suppl 08DEC2015 3 and osteo-differentiated H1-MP cells at D21). This thorough procedure still allowed inclusion of between 300 and nearly 2000 force versus indentation curves in the analysis for each cell type (Figs 1 and S1). 5 Microchannel separation device (MSD) The Microchannel Separation Device (MSD) was fabricated in a 1 mm thick fused silica substrate by femtosecond laser direct writing, followed by selective chemical etching. Laser writing of the MSD was performed using 390 fs pulses delivered from a commercial Yb-doped master oscillator power amplifier laser system (IMRA America FCPA μJewel D400) at a 10 repetition rate of 500 kHz and a central wavelength of 1047 ± 10 nm. The pulses were focused inside the volume of the fused silica substrate using a 0.4 NA (x20) aspheric objective lens. The translation speeds used were 4x10-3 ms-1 for the inlet ports, 2x10-3 ms-1 for the parallel microchannels and 0.5x10-3 ms-1 for the constrictions. The laser written device was subsequently etched using a 5% aqueous hydrofluoric acid solution for 1.5 hours. The total fabrication time 15 was 2.5 hours per device. The MSD has a footprint of 1.5 mm x 2.5 mm (Fig. S2A) and features four lateral fluid inlet ports (Fig. S2B) with a cross-section of 400 m x 400 m and a length of 600 m. The filter zone of the device consists of two parallel microfluidic channels (Fig. S2C) with a cross-section of 50 m x 50 m and a length of 500 m. The two microchannels are linked using 50 m long 20 constrictions with an average cross-section of 4 mm x 8 mm. The flow of cells and liquid within the MSD is supplied using a pair of software-controlled microfluidic pumps arranged to provide flow in either direction to both channels. This flow flexibility allowed pressure and flow-based Willoughby et al Suppl 08DEC2015 4 driving forces across the constrictions to be varied to achieve optimal performance. The size of the constriction channels was informed by CEM data and average cell size of human stem cells. 5 Figure Legends Fig. S1. CEM probability distributions of H1-MPs undergoing osteogenesis. For each of five time points (days 1, 6, 11, 15 and 21) during osteogenic differentiation of hESC-MP cells, , along with day 0 non-differentiated hESC-MP cells plated on HA as control, the discrete 10 probability distribution and resulting continuous probability density distribution for cell stiffness are shown (with Probability on the Y-axis in P(E) and CEM on the X-axis in kPa). The table summarizes the number of measurements and median CEM at each time point. Fig. S2. Microchannel Separation Device (MSD) design. (A) Four separation devices 15 photographed next to a British 5 pence coin for size comparison. (B-C) Optical microscope images providing (B) a top-view of the device showing inlet/outlet design, and (C) a close-up of the cross-flow design, with two parallel microfluidic channels linked using constrictions forming the filter zone. Willoughby et al Suppl 08DEC2015 5