Substrates panel
advertisement

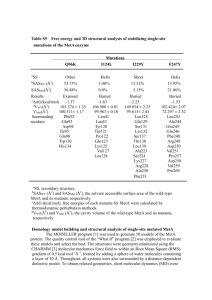
Supplementary Information Substrates panel The panel of substrates used for the screening was composed of glucose, galactose, fructose, mannose, xylose, arabinose, N-acetyl-D-glucosamine, sucrose, maltose, lactose, sorbitol, xylitol, cellobiose, cellotetraose, α-cyclodextrin, β-cyclodextrin, arabinan, chitosan, chitin, starch, maltotetraose, glycerol, hydroxymethylfurfural. Primers D279E_fw-5'GAACGGATACAGCGTCGAGGGTGCTTATATCGGTG3’ D279E_rev-5'CACCGATATAAGCACCCTCGACGCTGTATCCGTTC3' D279S_fw-5'-CTAAGAACGGATACAGCGTCAGTGGTGCTTATATCGGTGATG-3' D279S_rev-5'-CATCACCGATATAAGCACCACTGACGCTGTATCCGTTCTTAG3´ D337E_fw-5'-CATACGACGCACATGAGAACTTCTACGCCAGCA-3' D337E_rev-5'-TGCTGGCGTAGAAGTTCTCATGTGCGTCGTATG-3' G270E_fw-5'-GAGATCACCATGCAGATGGAGGTCTCTAAGAACGGATACA-3' G270E_rev-5'-TGTATCCGTTCTTAGAGACCTCCATCTGCATGGTGATCTC-3' G321Y_fw-5'-CTTGTTACTCACTTCGCCTATGCTGGCGTCAACGTCAA-3' G321Y_rev-5'-TTGACGTTGACGCCAGCATAGGCGAAGTGAGTAACAAG-3' G321N_fw-5'-CTTGTTACTCACTTCGCCAATGCTGGCGTCAACGTCAA-3' G321N_rev-5'-TTGACGTTGACGCCAGCATTGGCGAAGTGAGTAACAAG-3' M170Y_fw-5'-CACGGAGGCTACGGATATGTCGCCCGCAAGCAC-3' M170Y_rev-5'-GTGCTTGCGGGCGACATATCCGTAGCCTCCGTG-3' S410R_fw-5'-CAATTCTACGACAGGGTTGCCGCCACTGC-3' S410R_rev-5'-GCAGTGGCGGCAACCCTGTCGTAGAATTG-3' F319S_fw-5'-GGTCTTGTTACTCACAGCGCCGGTGCTGGCG-3' F319S_rev-5'-CGCCAGCACCGGCGCTGTGAGTAACAAGACC-3' W373F_fw-5'-GCAGCAGCCACTCTTGGTTCTTGCAAATGGACATCACC-3' W373F_rev-5'-GGTGATGTCCATTTGCAAGAACCAAGAGTGGCTGCTGC-3' W373Y_fw-5'-GCAGCAGCCACTCTTGGTATTTGCAAATGGACATCACC-3' W373Y_rev-5'-GGTGATGTCCATTTGCAAATACCAAGAGTGGCTGCTGC-3' L403W_fw-5'-GTCCACCGTGACACCCTCTGGCTCTTCCAATTCTACGACA-3' L403W_rev-5'-TGTCGTAGAATTGGAAGAGCCAGAGGGTGTCACGGTGGAC-3' The double mutant ChitO Q268R/S410R was constructed using the wild-type pET SUMO/chito as a template and both primers of Q268R and S410R. The previously constructed primer for Q268R by (Heuts et al., 2007) was obtained and the primer sequences are Q268R_fw 5´CCTGCCGAGATCACCATGCGCATGGGTGTCTCTAAGAACG-3´ and Q268R_rev 5´CGTTCTTAGAGACACCCATGCGCATGGTGATCTCGGCAGG-3´. For the mutant Q268R/G270E and Q268R/G270E/S410R an additional primer for Q268R/G270E was designed since the two mutation sites Gln268 and Gly270 are too close to each other. The sequence of the primers are Q268R/G270E_fw-5'-CTGCCGAGATCACCATGCGGATGGAG GTCTCTAAGAACGGATAC-3' and Q268R/G270E_rev-5'-GTATCCGTTCTTAGAGACCTCC ATCCGCATGGTGATCTC-3'. ChitO Q268R/G270E and the triple mutant Q268R/G270E/S410R were constructed with wild-type pET SUMO-chito and pET SUMO-chito S410R as a template respectively and the primers for Q268R/G270E were used. Steady state kinetic plots for substrate-ChitO mutant combinations that display substrate inhibition Fig. S1. Steady-state kinetic data of ChitO mutants that displayed substrate inhibition. Some mutants of ChitO showed different degrees of substrate inhibition as can be seen in the plots above. Fitting of data was performed using the provided equation for substrate inhibition (See Material and Methods). Flavin absorbance spectra absorbance 0.6 0.4 0.2 0.0 400 500 600 wavelength (nm) Fig. S2. Comparison of flavin absorbance spectrum of wild-type ChitO (38.8 µM, taken from Heuts et al. 2008) with the triple mutant ChitO Q268R/G270E/S410R (normalized for 38.8 µM). The relatively high absorbance below 400 nm is due to the presence of the 4a-spirohydantoin FAD degradation product formed during the purification process as has also been observed for sequencerelated the berberine bridge enzyme (Winkler et al., 2008). The content of this inactivated FAD derivate in the ChitO preparations differed from batch to batch for wild-type and mutant enzymes. However, enzyme concentrations were estimated based on the known extinction coefficient for the oxidized FAD cofactor of wild-type ChitO at 445 nm, therefore only the concentration of intact FAD, thus active enzyme, was used to determine the turnover rates. References - - Heuts DPHM, Janssen DB, Fraaije MW. 2007. Changing the substrate specificity of a chitooligosaccharide oxidase from Fusarium graminearum by model-inspired site-directed mutagenesis. FEBS Lett. 581:4905–9. Winkler A, Lyskowski A, Riedl S, Puhl M, Kutchan TM, Macheroux P, Gruber K. 2008. A concerted mechanism for berberine bridge enzyme. Nat. Chem. Biol., 12: 739–41.