click here
advertisement

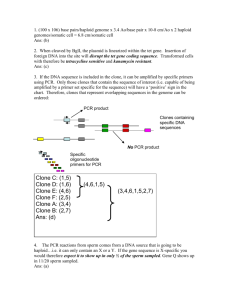
Answers to Quiz 4: The number of codons that can specify each individual amino acid is indicated in above the peptide sequence: 2 6 2 2 1 3 1 4 1 6 1 4 2 2 1 Phe-Ser-Glu-Tyr-Met-Ile-Trp-Val-Met-Arg-Tyr-Ala-Gln-Tyr-Met a) Phe-Ser-Glu-Tyr-Met-Ile-Trp (144); b) Met-Ile-TrpVal-Met-Arg-Tyr (72); c) Glu-Tyr-Met-Ile-Trp-Val-Met (48); d) Met-Arg-Tyr-Ala-Gln-Tyr-Met (96). 1. The lowest redundancy is found in Glu-Tyr-Met-IleTrp-Val-Met. Ans: (c). 2. This peptide could be encoded by 48 different codon sequences. Ans: (b). 3. cDNA libraries are derived from mRNAs, enzymatically converted to DNA using reverse transcriptase and DNA polymerase. Ans: (c). 4. If we built a probe that was based on the codon sequence, it would be a ‘sense’ probe with the same nucleotide sequence as the messenger RNA. It therefore would not have sequence complementarity to the mRNA, and couldn’t bind to mRNA in a Northern blot. Ans: True. 5. Individual I-1 is heterozygous for two different RFLP patterns; one of his homologs contains a 7 kb fragment that hybridizes with the probe, and the other contains a restriction site that allows the probe to hybridize to a 4 and 3 kb piece: e.g. 7.0 kb 7 I-1 4 * 3 5.3 kb 4.0 kb 3.0 kb 1.7 kb Individual I-2 also contains one homolog that hybridizes to a Piece. In this individual, however, the 7 kb fragment that hybridizes to the probe looks like it can be cleaved into two fragments due to the presence of a variable internal restriction site that cleaves the fragment into a 5.3 and 1.7 kb piece: e.g. 7 kb. 7 I-2 5.3 1.7 * Following the segregation of both patterns through the pedigree, it seems as if the 7 kb fragment donated by I-2 is always seen when the disease is present, and the 5.3/1.7 pattern is always seen when the disease is absent, with the exception of individual II-7. This can be explained if the disease gene locus is linked to the chromosome carrying the 7 kb fragment. Individual II-7 can be explained by a crossover occurring during meiosis in individual I-2, that occurs between the RFLP locus and the disease gene locus: e.g. 7 5.3 1.7 D X + 5. The 7 kb fragment is linked to the disease gene. Ans: (d). 6. The distance between the RFLP locus and the disease gene is 1/13 or between 7-8 map units. Ans: (c). 7. The DNA sequence for the STRAND BEING SYNTHESIZED would read from the bottom of the gel: 5’-CGTGCACGTGCA-3’ The DNA sequence for the TEMPLATE STRAND would be complementary and run antiparallel to this sequence, i.e. 5’-CGTGCACGTGCA-3’ 3’-GCACGTGCACGT-5’ Ans: (c). 8. All of the sequencing reactions need to contain all of the normal components for DNA synthesis, and deoxyadenosine triphosphate (dATP) is a normal component of DNA. The individual reactions only contain one chain terminator- a dideoxynucleotide that contains neither a 3’ or 5’ hydroxyl group on the sugar. So dideoxyATP- ddATP would be present in the ‘A’ reaction, ddTTP in the ‘T’ reaction, etc. terminating at specific bases as the template strand is being read. Ans: False. The pathway would be: W 6 R 2 V 3 U 4 S 5 T 1 Q 9. The immediate precursor to S would be U. Ans: (d). 10. A mutation in gene 3 would lead to accumulation of precursor V. Ans: (e).